Richard J. Chen

I am a 5th year Ph.D. Candidate (and NSF-GRFP Fellow) advised by Faisal Mahmood at Harvard University, and also within Brigham and Women’s Hospital, Dana-Farber Cancer Institute, and the Broad Institute.
Prior to starting my Ph.D., I obtained my B.S/M.S. in Biomedical Engineering and Computer Science at Johns Hopkins University, where I worked with Nicholas Durr and Alan Yuille. In industry, I have also worked at Apple Inc. in the Health Special Project and Applied Machine Learning Groups (with Belle Tseng and Andrew Trister), and at Microsoft Research in the BioML Group (with Rahul Gopalkrishnan).
Research Highlights
-
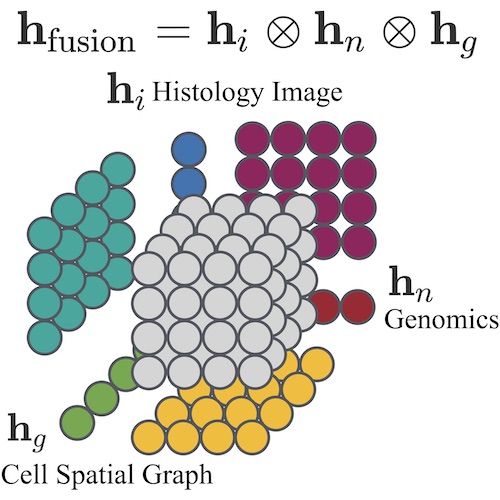
Multimodal Integration: Multimodal learning has emerged as an interdisciplinary field to solve many core problems in machine perception, human-computer interaction, and recently in biology & medicine, in which there is often an enormous wealth of multimodal data collected in parallel to study the same underlying disease. I have worked on a range of problems in multimodal learning for integrating: 1) multimodal sensor streams from the Apple Watch and iPhone Data to predict mild cognitive decline, 2) RGB and depth images for non-polyploidal lesion classification and SLAM in surgical robotics, and 3) pathology images and genomics for cancer prognosis.
-
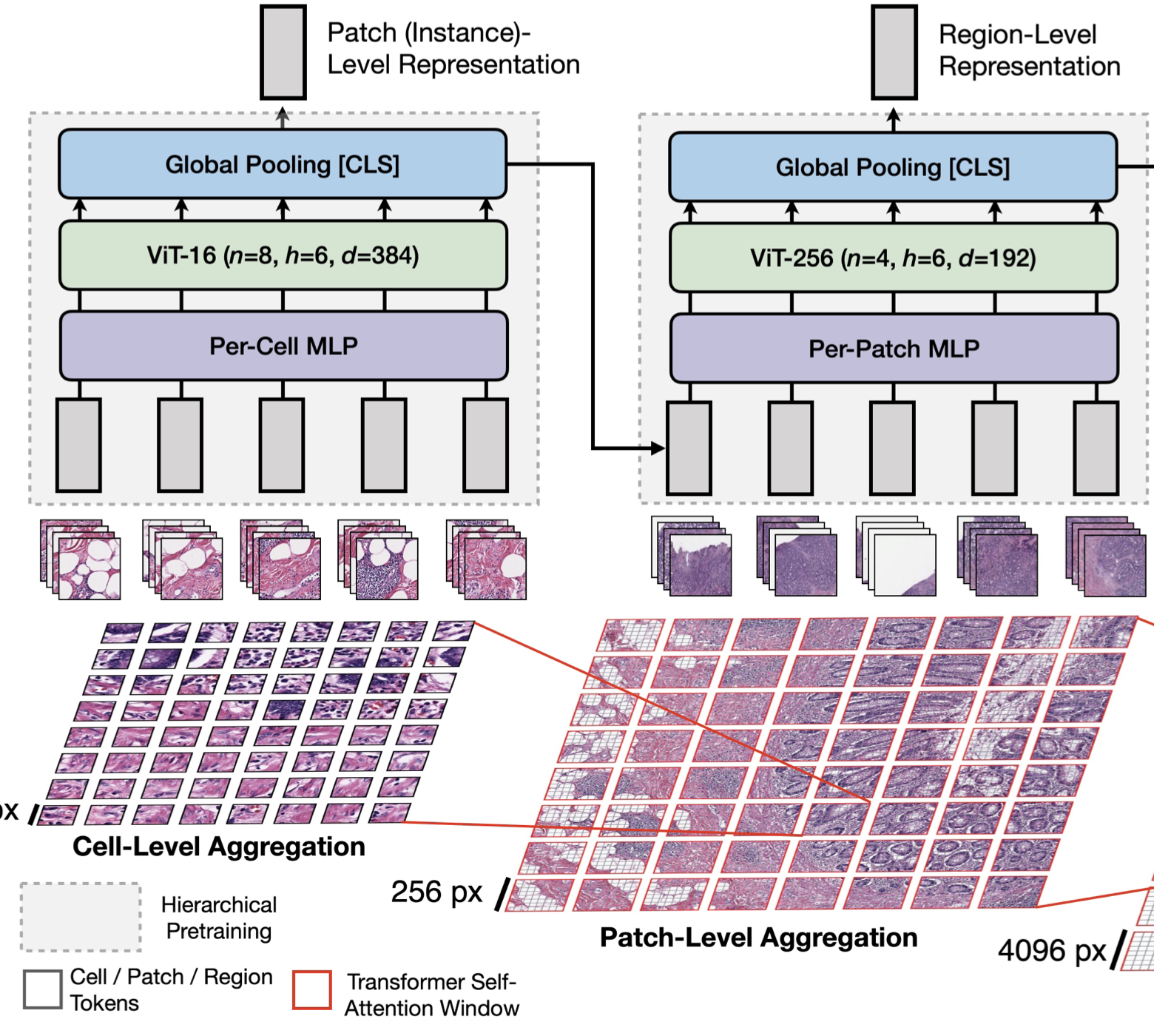
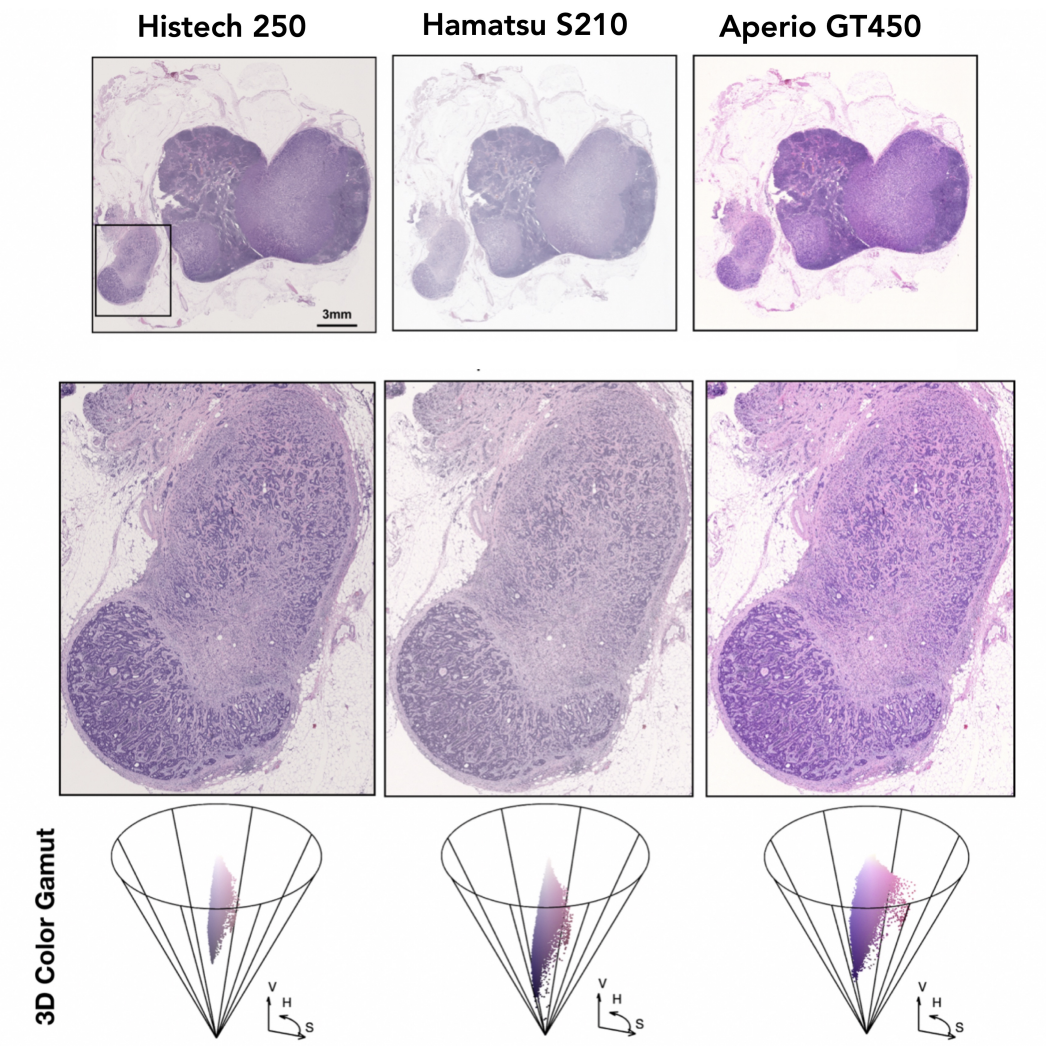
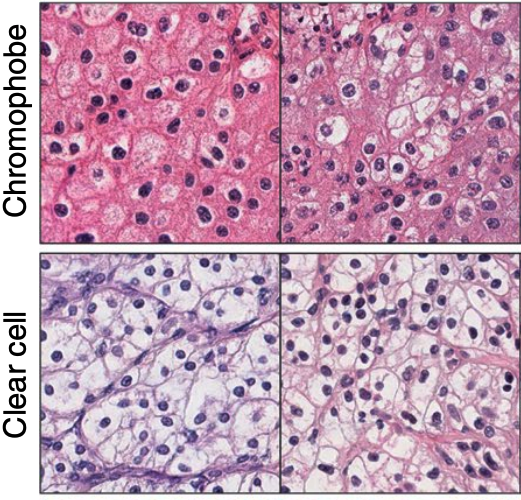
Representation Learning for Gigapixel Images: Though deep learning has revolutionized computer vision in many disciplines, gigapixel whole-slide imaging (WSI) in computational pathology is a complex computer vision domain that renders traditional, Convnet-based supervised learning approaches infeasible. To address this issue, I have been working on interpreting large gigapixel images as permutation-invariant sets (or bags in MIL literature), and then developing Transformer-based approaches for weakly-supervised learning and self-supervised learning in WSIs.
-
Generative AI & Healthcare Policy: “What constitutes authenticity, and how would the lack of authenticity shape our perception of reality?” The science fiction American writer Philip K. Dick posited similar questions throughout his literary career and, in particular, in his 1972 essay “How to build a universe that doesn’t fall apart two days later”. I am interested in: using synthetic data for domain adaptation / generalization, developing synthetic environments for simulating challenging scenarios for neural networks, as well as the the policy challenges in training AI-SaMDs with synthetic data,
Recent News
| Aug, 2023 | Excited to share our latest preprint on UNI, a general-purpose self-supervised model for computational pathology. In addition, my Master’s student, Tong Ding, is joining the Computer Science Ph.D. program at Harvard University (SEAS). Congratulations Tong! |
|---|---|
| Jul, 2023 | Our perspective on algorithm fairness in AI and medicine/healthcare was published in Nature BME. In addition, excited to share our latest preprint on CONCH (CONtrastive learning from Captions for Histopathology), a visual-language foundation model for computational pathology. Stay tuned! |
| Jun, 2023 | Our work on zero-shot slide classification with visual-language pretraining was published in CVPR. Code + pretrained model weights are made available. |
| Aug, 2022 | Our work on PORPOISE (Pathology-Omic Research Platform for Integrated Survival Estimation), and our review on multimodal learning for oncology were both published in Cancer Cell. See the associated demo! |
| Jun, 2022 | Our work on Hierarchical Image Pyramid Transformer (HIPT) is highlighted as an Oral Presentation in CVPR, and as a Spotlight Talk in the Transformers 4 Vision (T4V) CVPR Workshop. Code + pretrained model weights are made available. Lastly, my visiting student, Yicong Li, is joining the Computer Science Ph.D. program at Harvard University (SEAS). Congratulations Yicong! |
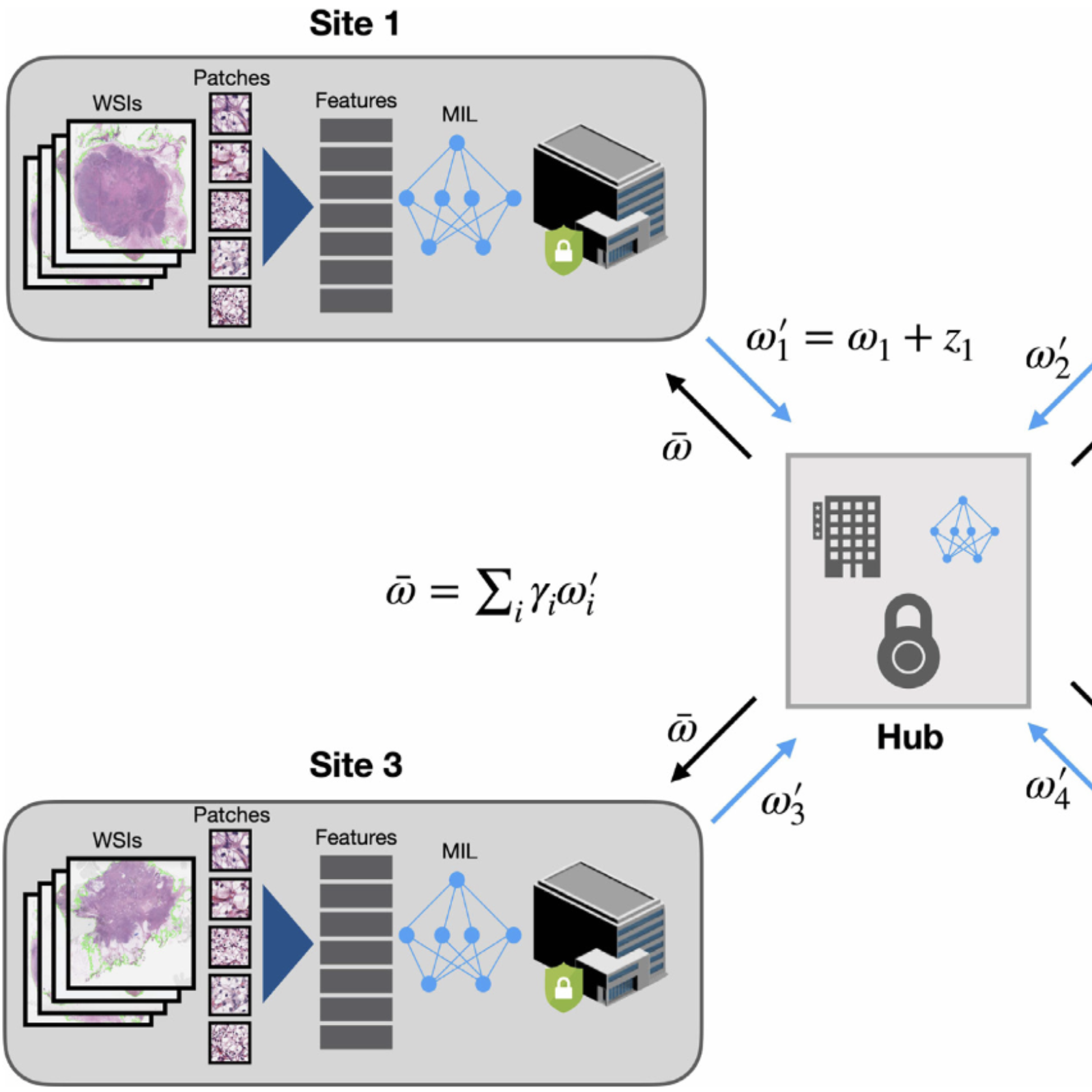
| Mar, 2022 | Our work on CRANE was published in Nature Medicine. Also, code + pretrained model weights are made available for our recent Self-Supervised ViT work in NeurIPSW LMRL 2021. Lastly, our work on federated learning for CPATH (HistoFL) was published in Medical Image Analysis. |
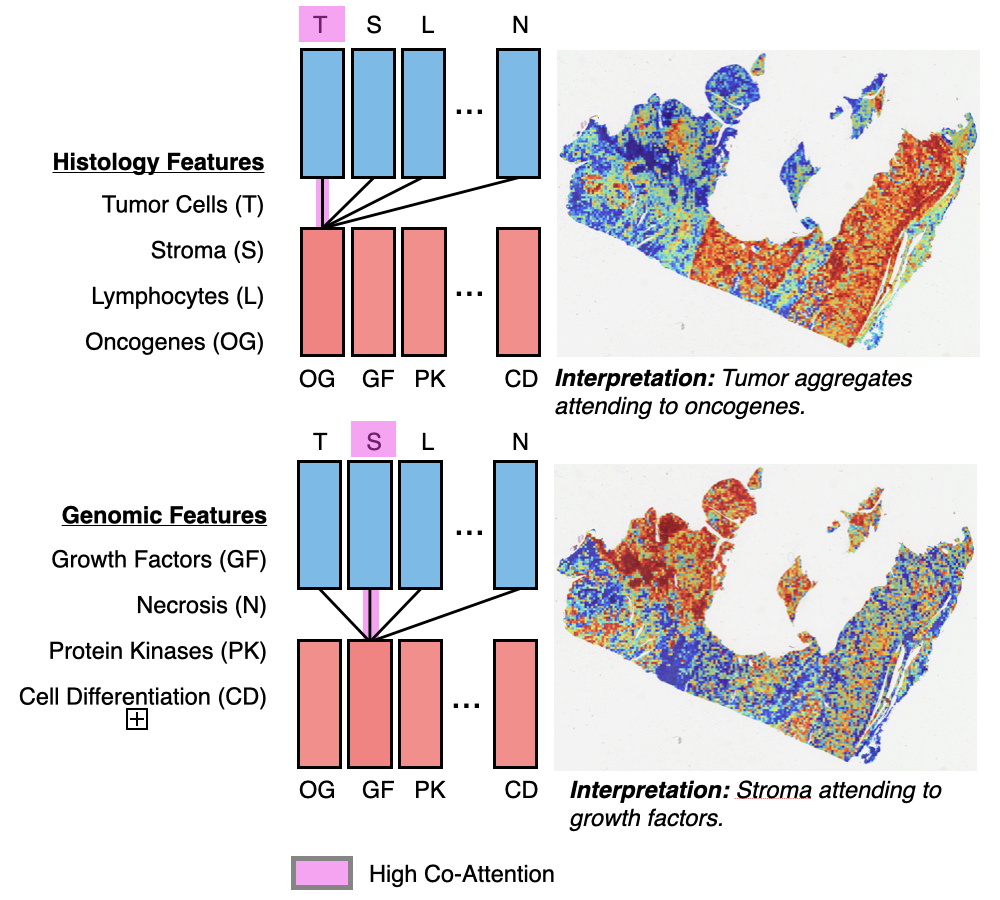
| Jul, 2021 | Joined Microsoft Research as an PhD Research Intern, working with Rahul Gopalkrishnan in the BioML Group. In press, our commentary on synthetic data for machine learning and healthcare was also published in Nature BME. Lastly, two papers, Patch-GCN and Multimodal Co-Attention Transformers (MCAT), were accepted into MICCAI and ICCV respectively. |