Publications
2023
 A General-Purpose Self-Supervised Model for Computational PathologyRichard J. Chen, Tong Ding, Ming Y. Lu, Drew F. K. Williamson, Guillaume Jaume, Bowen Chen, Andrew Zhang, Daniel Shao, Andrew H. Song, Muhammad Shaban, Mane Williams, Anurag Vaidya, Sharifa Sahai, Lukas Oldenburg, Luca L. Weishaupt, Judy J. Wang, Walt Williams, Long Phi Le, Georg Gerber, and Faisal MahmoodarXiv preprint arXiv:TBD 2023
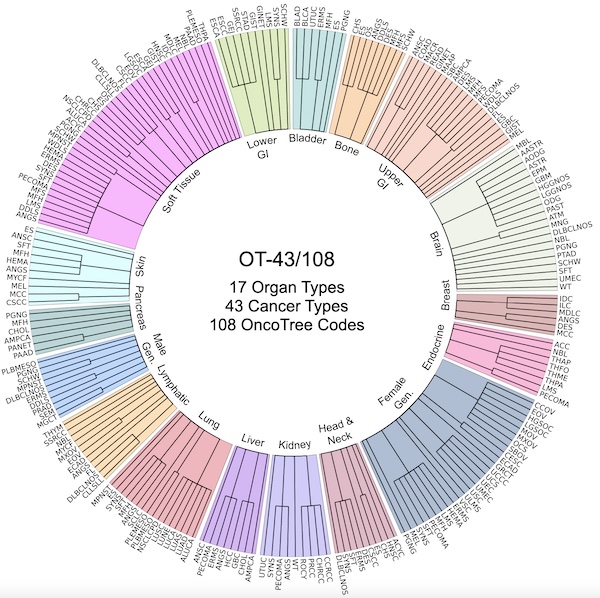
A General-Purpose Self-Supervised Model for Computational PathologyRichard J. Chen, Tong Ding, Ming Y. Lu, Drew F. K. Williamson, Guillaume Jaume, Bowen Chen, Andrew Zhang, Daniel Shao, Andrew H. Song, Muhammad Shaban, Mane Williams, Anurag Vaidya, Sharifa Sahai, Lukas Oldenburg, Luca L. Weishaupt, Judy J. Wang, Walt Williams, Long Phi Le, Georg Gerber, and Faisal MahmoodarXiv preprint arXiv:TBD 2023Tissue phenotyping is a fundamental computational pathology (CPath) task in learning objective characterizations of histopathologic biomarkers in anatomic pathology. However, whole-slide imaging (WSI) poses a complex computer vision problem in which the large-scale image resolutions of WSIs and the enormous diversity of morphological phenotypes preclude large-scale data annotation. Current efforts have proposed using pretrained image encoders with either transfer learning from natural image datasets or self-supervised pretraining on publicly-available histopathology datasets, but have not been extensively developed and evaluated across diverse tissue types at scale. We introduce UNI, a general-purpose, self-supervised model for pathology, pretrained using over 100 million tissue patches from over 100,000 diagnostic haematoxylin and eosin-stained WSIs across 20 major tissue types, and evaluated on 33 representative CPath clinical tasks in CPath of varying diagnostic difficulties. In addition to outperforming previous state-of-the-art models, we demonstrate new modeling capabilities in CPath such as resolution-agnostic tissue classification, slide classification using few-shot class prototypes, and disease subtyping generalization in classifying up to 108 cancer types in the OncoTree code classification system. UNI advances unsupervised representation learning at scale in CPath in terms of both pretraining data and downstream evaluation, enabling data-efficient AI models that can generalize and transfer to a gamut of diagnostically-challenging tasks and clinical workflows in anatomic pathology.
@article{chen2023general, title = {A General-Purpose Self-Supervised Model for Computational Pathology}, author = {Chen, Richard J. and Ding, Tong and Lu, Ming Y. and Williamson, Drew F. K. and Jaume, Guillaume and Chen, Bowen and Zhang, Andrew and Shao, Daniel and Song, Andrew H. and Shaban, Muhammad and Williams, Mane and Vaidya, Anurag and Sahai, Sharifa and Oldenburg, Lukas and Weishaupt, Luca L. and Wang, Judy J. and Williams, Walt and Le, Long Phi and Gerber, Georg and Mahmood, Faisal}, journal = {arXiv preprint arXiv:TBD}, year = {2023}, arxiv = {TBD}, abbr = {chen2023general.jpg}, selected = {true} }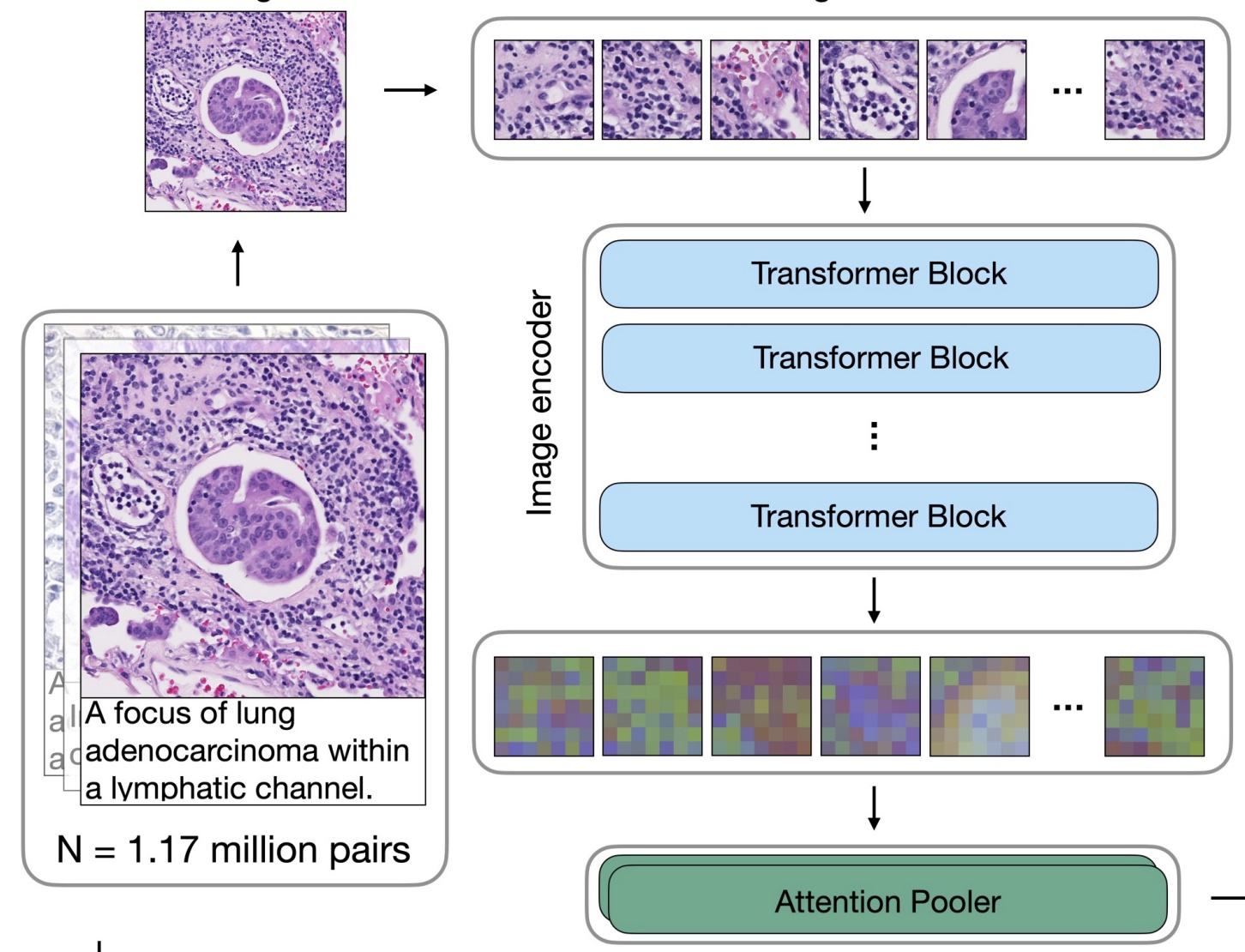 Towards a Visual-Language Foundation Model for Computational PathologyMing Y Lu, Bowen Chen, Drew FK Williamson, Richard J Chen, Ivy Liang, Tong Ding, Guillaume Jaume, Igor Odintsov, Andrew Zhang, Long Phi Le, and Faisal MahmoodarXiv preprint arXiv:2307.12914 2023
Towards a Visual-Language Foundation Model for Computational PathologyMing Y Lu, Bowen Chen, Drew FK Williamson, Richard J Chen, Ivy Liang, Tong Ding, Guillaume Jaume, Igor Odintsov, Andrew Zhang, Long Phi Le, and Faisal MahmoodarXiv preprint arXiv:2307.12914 2023The accelerated adoption of digital pathology and advances in deep learning have enabled the development of powerful models for various pathology tasks across a diverse array of diseases and patient cohorts. However, model training is often difficult due to label scarcity in the medical domain and the model’s usage is limited by the specific task and disease for which it is trained. Additionally, most models in histopathology leverage only image data, a stark contrast to how humans teach each other and reason about histopathologic entities. We introduce CONtrastive learning from Captions for Histopathology (CONCH), a visual-language foundation model developed using diverse sources of histopathology images, biomedical text, and notably over 1.17 million image-caption pairs via task-agnostic pretraining. Evaluated on a suite of 13 diverse benchmarks, CONCH can be transferred to a wide range of downstream tasks involving either or both histopathology images and text, achieving state-of-the-art performance on histology image classification, segmentation, captioning, text-to-image and image-to-text retrieval. CONCH represents a substantial leap over concurrent visual-language pretrained systems for histopathology, with the potential to directly facilitate a wide array of machine learning-based workflows requiring minimal or no further supervised fine-tuning.
@article{lu2023towards, title = {Towards a Visual-Language Foundation Model for Computational Pathology}, author = {Lu, Ming Y and Chen, Bowen and Williamson, Drew FK and Chen, Richard J and Liang, Ivy and Ding, Tong and Jaume, Guillaume and Odintsov, Igor and Zhang, Andrew and Le, Long Phi and Mahmood, Faisal}, journal = {arXiv preprint arXiv:2307.12914}, year = {2023}, arxiv = {2307.12914}, abbr = {lu2023towards.jpg} }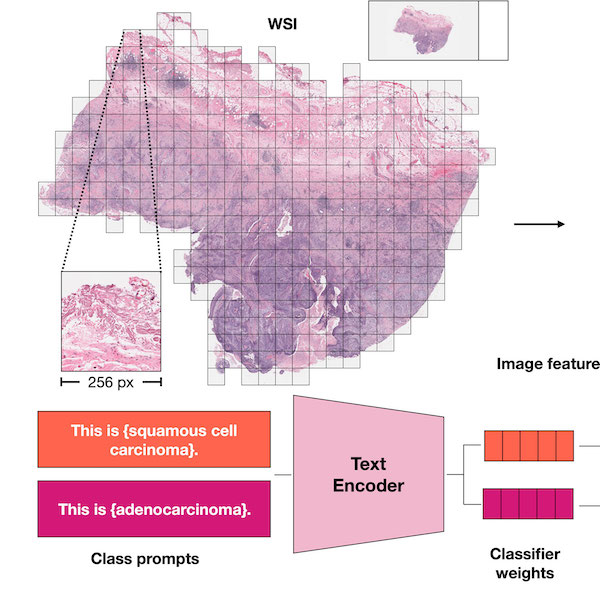 Visual Language Pretrained Multiple Instance Zero-Shot Transfer for Histopathology ImagesMing Y. Lu, Bowen Chen, Andrew Zhang, Drew F. K. Williamson, Richard J. Chen, Tong Ding, Long Phi Le, Yung-Sung Chuang, and Faisal MahmoodIn Proceedings of the IEEE/CVF Conference on Computer Vision and Pattern Recognition (CVPR) 2023
Visual Language Pretrained Multiple Instance Zero-Shot Transfer for Histopathology ImagesMing Y. Lu, Bowen Chen, Andrew Zhang, Drew F. K. Williamson, Richard J. Chen, Tong Ding, Long Phi Le, Yung-Sung Chuang, and Faisal MahmoodIn Proceedings of the IEEE/CVF Conference on Computer Vision and Pattern Recognition (CVPR) 2023Contrastive visual language pretraining has emerged as a powerful method for either training new language-aware image encoders or augmenting existing pretrained models with zero-shot visual recognition capabilities. However, existing works typically train on large datasets of image-text pairs and have been designed to perform downstream tasks involving only small to medium sized-images, neither of which are applicable to the emerging field of computational pathology where there are limited publicly available paired image-text datasets and each image can span up to 100,000 x 100,000 pixels in dimensions. In this paper we present MI-Zero, a simple and intuitive framework for unleashing the zero-shot transfer capabilities of contrastively aligned image and text models to gigapixel histopathology whole slide images, enabling multiple downstream diagnostic tasks to be carried out by pretrained encoders without requiring any additional labels. MI-Zero reformulates zero-shot transfer under the framework of multiple instance learning to overcome the computational challenge of inference on extremely large images. We used over 550k pathology reports and other available in-domain text corpora to pretrain our text encoder. By effectively leveraging strong pretrained encoders, our best model pretrained on over 33k histopathology image-caption pairs achieves an average median zero-shot accuracy of 70.2% across three different real-world cancer subtyping tasks. Our code is available at: https://github.com/mahmoodlab/MI-Zero.
@inproceedings{lu2023visual, author = {Lu, Ming Y. and Chen, Bowen and Zhang, Andrew and Williamson, Drew F. K. and Chen, Richard J. and Ding, Tong and Le, Long Phi and Chuang, Yung-Sung and Mahmood, Faisal}, title = {Visual Language Pretrained Multiple Instance Zero-Shot Transfer for Histopathology Images}, booktitle = {Proceedings of the IEEE/CVF Conference on Computer Vision and Pattern Recognition (CVPR)}, month = jun, year = {2023}, pages = {19764-19775}, url = {https://openaccess.thecvf.com/content/CVPR2023/html/Lu_Visual_Language_Pretrained_Multiple_Instance_Zero-Shot_Transfer_for_Histopathology_Images_CVPR_2023_paper.html}, code = {https://github.com/mahmoodlab/MI-Zero}, abbr = {lu2023mizero.jpeg} } Algorithmic fairness in artificial intelligence for medicine and healthcareRichard J. Chen, Judy J. Wang, Drew FK. Williamson, Tiffany Y. Chen, Jana Lipkova, Ming Y. Lu, Sharifa Sahai, and Faisal MahmoodNature Biomedical Engineering 2023
Algorithmic fairness in artificial intelligence for medicine and healthcareRichard J. Chen, Judy J. Wang, Drew FK. Williamson, Tiffany Y. Chen, Jana Lipkova, Ming Y. Lu, Sharifa Sahai, and Faisal MahmoodNature Biomedical Engineering 2023In healthcare, the development and deployment of insufficiently fair systems of artificial intelligence (AI) can undermine the delivery of equitable care. Assessments of AI models stratified across subpopulations have revealed inequalities in how patients are diagnosed, treated and billed. In this Perspective, we outline fairness in machine learning through the lens of healthcare, and discuss how algorithmic biases (in data acquisition, genetic variation and intra-observer labelling variability, in particular) arise in clinical workflows and the resulting healthcare disparities. We also review emerging technology for mitigating biases via disentanglement, federated learning and model explainability, and their role in the development of AI-based software as a medical device.
@article{chen2023algorithmic, title = {Algorithmic fairness in artificial intelligence for medicine and healthcare}, author = {Chen, Richard J. and Wang, Judy J. and Williamson, Drew FK. and Chen, Tiffany Y. and Lipkova, Jana and Lu, Ming Y. and Sahai, Sharifa and Mahmood, Faisal}, journal = {Nature Biomedical Engineering}, volume = {7}, number = {6}, pages = {719--742}, year = {2023}, publisher = {Nature Publishing Group UK London}, selected = {true}, abbr = {chen2021algorithm.png}, url = {https://www.nature.com/articles/s41551-023-01056-8} }
2022
 Artificial intelligence for multimodal data integration in oncologyJana Lipkova, Richard J Chen, Bowen Chen, Ming Y Lu, Matteo Barbieri, Daniel Shao, Anurag J Vaidya, Chengkuan Chen, Luoting Zhuang, Drew FK Williamson, and othersCancer Cell 2022
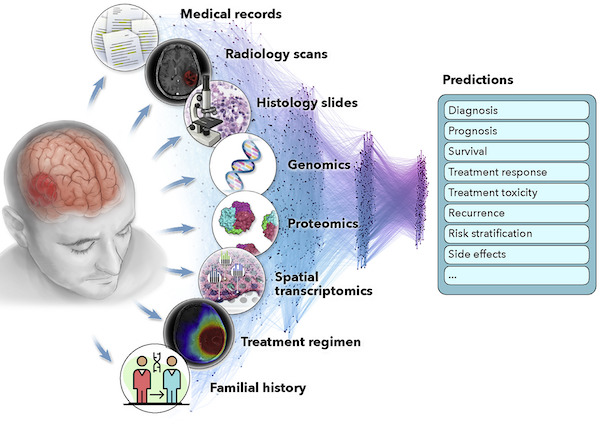
Artificial intelligence for multimodal data integration in oncologyJana Lipkova, Richard J Chen, Bowen Chen, Ming Y Lu, Matteo Barbieri, Daniel Shao, Anurag J Vaidya, Chengkuan Chen, Luoting Zhuang, Drew FK Williamson, and othersCancer Cell 2022@article{lipkova2022artificial, title = {Artificial intelligence for multimodal data integration in oncology}, author = {Lipkova, Jana and Chen, Richard J and Chen, Bowen and Lu, Ming Y and Barbieri, Matteo and Shao, Daniel and Vaidya, Anurag J and Chen, Chengkuan and Zhuang, Luoting and Williamson, Drew FK and others}, journal = {Cancer Cell}, volume = {40}, number = {10}, pages = {1095--1110}, year = {2022}, publisher = {Elsevier}, url = {https://www.cell.com/cancer-cell/fulltext/S1535-6108(22)00441-X}, abbr = {lipkova2022art.jpg} }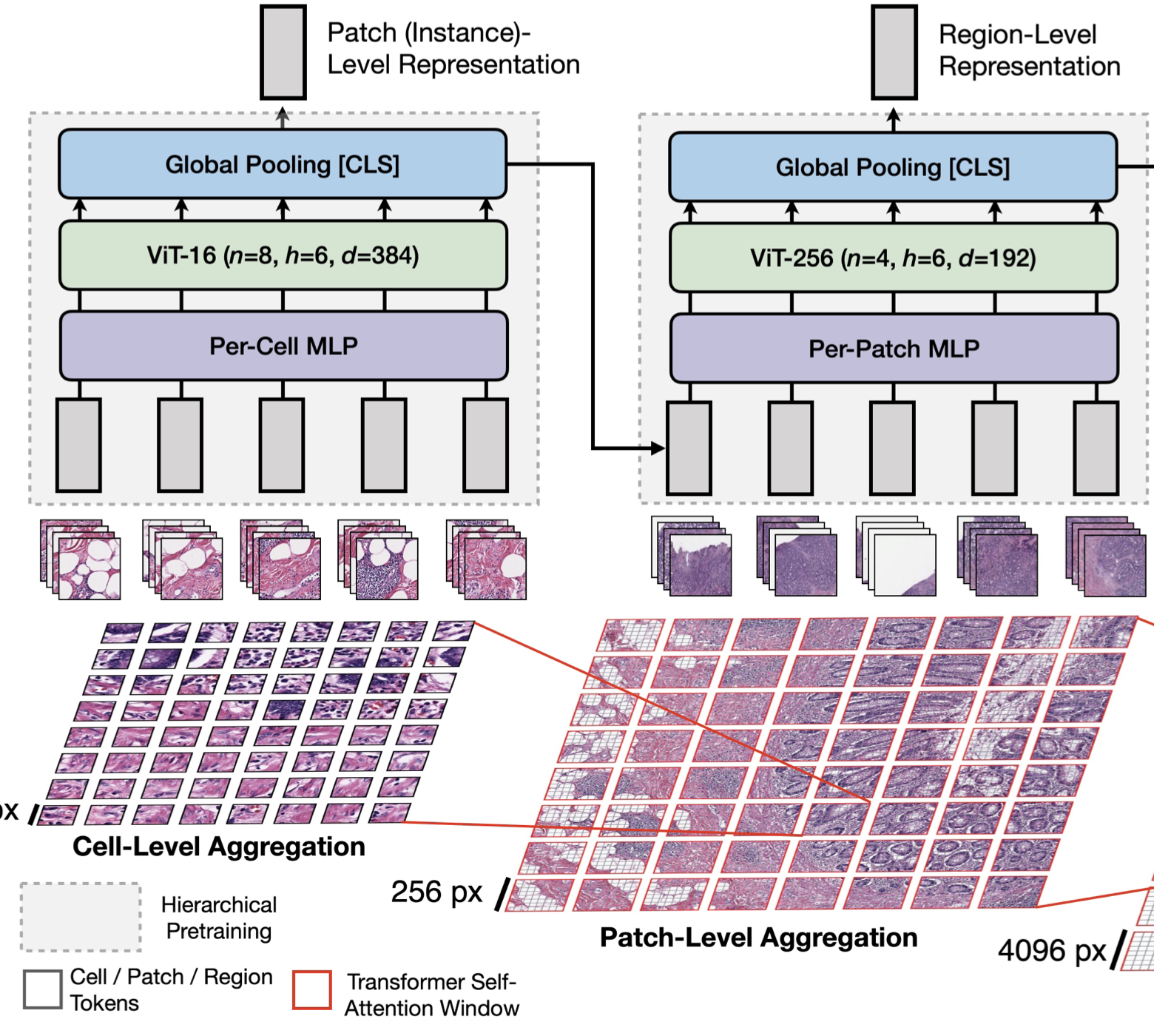 Scaling Vision Transformers to Gigapixel Images via Hierarchical Self-Supervised LearningRichard J Chen, Chengkuan Chen, Yicong Li, Tiffany Y Chen, Andrew D Trister, Rahul G Krishnan, and Faisal MahmoodIn Proceedings of the IEEE/CVF Conference on Computer Vision and Pattern Recognition 2022Oral Presentation
Scaling Vision Transformers to Gigapixel Images via Hierarchical Self-Supervised LearningRichard J Chen, Chengkuan Chen, Yicong Li, Tiffany Y Chen, Andrew D Trister, Rahul G Krishnan, and Faisal MahmoodIn Proceedings of the IEEE/CVF Conference on Computer Vision and Pattern Recognition 2022Oral PresentationVision Transformers (ViTs) and their multi-scale and hierarchical variations have been successful at capturing image representations but their use has been generally studied for low-resolution images (e.g. - 256x256, 384x384). For gigapixel whole-slide imaging (WSI) in computational pathology, WSIs can be as large as 150000x150000 pixels at 20x magnification and exhibit a hierarchical structure of visual tokens across varying resolutions: from 16x16 images capture spatial patterns among cells, to 4096x4096 images characterizing interactions within the tissue microenvironment. We introduce a new ViT architecture called the Hierarchical Image Pyramid Transformer (HIPT), which leverages the natural hierarchical structure inherent in WSIs using two levels of self-supervised learning to learn high-resolution image representations. HIPT is pretrained across 33 cancer types using 10,678 gigapixel WSIs, 408,218 4096x4096 images, and 104M 256x256 images. We benchmark HIPT representations on 9 slide-level tasks, and demonstrate that: 1) HIPT with hierarchical pretraining outperforms current state-of-the-art methods for cancer subtyping and survival prediction, 2) self-supervised ViTs are able to model important inductive biases about the hierarchical structure of phenotypes in the tumor microenvironment.
@inproceedings{chen2022scaling, title = {Scaling Vision Transformers to Gigapixel Images via Hierarchical Self-Supervised Learning}, author = {Chen, Richard J and Chen, Chengkuan and Li, Yicong and Chen, Tiffany Y and Trister, Andrew D and Krishnan, Rahul G and Mahmood, Faisal}, booktitle = {Proceedings of the IEEE/CVF Conference on Computer Vision and Pattern Recognition}, year = {2022}, pages = {16144--16155}, code = {https://github.com/mahmoodlab/HIPT}, url = {https://openaccess.thecvf.com/content/CVPR2022/html/Chen_Scaling_Vision_Transformers_to_Gigapixel_Images_via_Hierarchical_Self-Supervised_Learning_CVPR_2022_paper.html}, arxiv = {2206.02647}, abbr = {chen2022scaling.png}, selected = {true}, honor = {Oral Presentation} }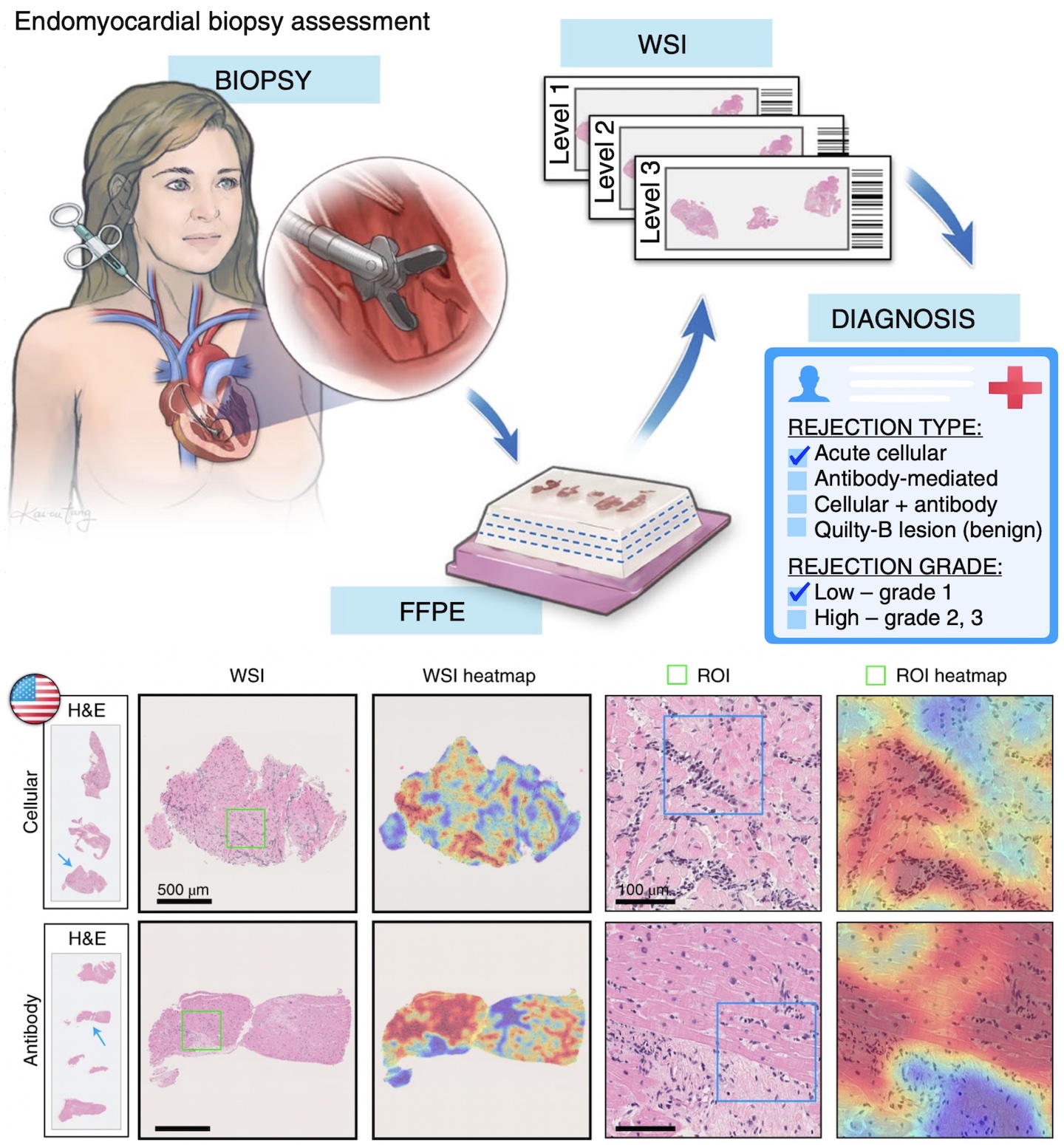 Deep Learning-Enabled Assessment of Cardiac Allograft Rejection from Endomyocardial BiopsiesJana Lipkova, Tiffany Y. Chen, Ming Y. Lu, Richard J. Chen, Maha Shady, Mane Williams, Jingwen Wang, Zahra Noor, Richard N. Mitchell, Mehmet Turan, Gulfize Coskun, Funda Yilmaz, Derya Demir, Deniz Nart, Kayhan Basak, Nesrin Turhan, Selvinaz Ozkara, Yara Banz, Katja E. Odenring, and Faisal MahmoodNature Medicine 2022
Deep Learning-Enabled Assessment of Cardiac Allograft Rejection from Endomyocardial BiopsiesJana Lipkova, Tiffany Y. Chen, Ming Y. Lu, Richard J. Chen, Maha Shady, Mane Williams, Jingwen Wang, Zahra Noor, Richard N. Mitchell, Mehmet Turan, Gulfize Coskun, Funda Yilmaz, Derya Demir, Deniz Nart, Kayhan Basak, Nesrin Turhan, Selvinaz Ozkara, Yara Banz, Katja E. Odenring, and Faisal MahmoodNature Medicine 2022Endomyocardial biopsy (EMB) screening represents the standard of care for detecting allograft rejections after heart transplant. Manual interpretation of EMBs is affected by substantial interobserver and intraobserver variability, which often leads to inappropriate treatment with immunosuppressive drugs, unnecessary follow-up biopsies and poor transplant outcomes. Here we present a deep learning-based artificial intelligence (AI) system for automated assessment of gigapixel whole-slide images obtained from EMBs, which simultaneously addresses detection, subtyping and grading of allograft rejection. To assess model performance, we curated a large dataset from the United States, as well as independent test cohorts from Turkey and Switzerland, which includes large-scale variability across populations, sample preparations and slide scanning instrumentation. The model detects allograft rejection with an area under the receiver operating characteristic curve (AUC) of 0.962; assesses the cellular and antibody-mediated rejection type with AUCs of 0.958 and 0.874, respectively; detects Quilty B lesions, benign mimics of rejection, with an AUC of 0.939; and differentiates between low-grade and high-grade rejections with an AUC of 0.833. In a human reader study, the AI system showed non-inferior performance to conventional assessment and reduced interobserver variability and assessment time. This robust evaluation of cardiac allograft rejection paves the way for clinical trials to establish the efficacy of AI-assisted EMB assessment and its potential for improving heart transplant outcomes.
@article{lipkova2022deep, doi = {10.1038/s41591-022-01709-2}, url = {https://doi.org/10.1038/s41591-022-01709-2}, year = {2022}, month = mar, publisher = {Springer Science and Business Media {LLC}}, volume = {28}, number = {3}, pages = {575-582}, author = {Lipkova, Jana and Chen, Tiffany Y. and Lu, Ming Y. and Chen, Richard J. and Shady, Maha and Williams, Mane and Wang, Jingwen and Noor, Zahra and Mitchell, Richard N. and Turan, Mehmet and Coskun, Gulfize and Yilmaz, Funda and Demir, Derya and Nart, Deniz and Basak, Kayhan and Turhan, Nesrin and Ozkara, Selvinaz and Banz, Yara and Odenring, Katja E. and Mahmood, Faisal}, title = {Deep Learning-Enabled Assessment of Cardiac Allograft Rejection from Endomyocardial Biopsies}, journal = {Nature Medicine}, demo = {http://crane.mahmoodlab.org}, abbr = {lipkova2022deep.png}, code = {https://github.com/mahmoodlab/CRANE}, press = {https://hms.harvard.edu/news/heart-saving-ai} } Pan-Cancer Integrative Histology-Genomic Analysis via Multimodal Deep LearningRichard J Chen, Ming Y Lu, Drew FK Williamson, Tiffany Y Chen, Jana Lipkova, Muhammad Shaban, Maha Shady, Mane Williams, Bumjin Joo, Zahra Noor, and Faisal MahmoodCancer Cell 2022Best Paper, Case Western Artificial Intelligence in Oncology Symposium, 2020. Cover Art of Cancer Cell (Volume 40 Issue 8).
Pan-Cancer Integrative Histology-Genomic Analysis via Multimodal Deep LearningRichard J Chen, Ming Y Lu, Drew FK Williamson, Tiffany Y Chen, Jana Lipkova, Muhammad Shaban, Maha Shady, Mane Williams, Bumjin Joo, Zahra Noor, and Faisal MahmoodCancer Cell 2022Best Paper, Case Western Artificial Intelligence in Oncology Symposium, 2020. Cover Art of Cancer Cell (Volume 40 Issue 8).Summary The rapidly emerging field of computational pathology has demonstrated promise in developing objective prognostic models from histology images. However, most prognostic models are either based on histology or genomics alone and do not address how these data sources can be integrated to develop joint image-omic prognostic models. Additionally, identifying explainable morphological and molecular descriptors from these models that govern such prognosis is of interest. We use multimodal deep learning to jointly examine pathology whole-slide images and molecular profile data from 14 cancer types. Our weakly supervised, multimodal deep-learning algorithm is able to fuse these heterogeneous modalities to predict outcomes and discover prognostic features that correlate with poor and favorable outcomes. We present all analyses for morphological and molecular correlates of patient prognosis across the 14 cancer types at both a disease and a patient level in an interactive open-access database to allow for further exploration, biomarker discovery, and feature assessment.
@article{chen2021pan, title = {Pan-Cancer Integrative Histology-Genomic Analysis via Multimodal Deep Learning}, journal = {Cancer Cell}, volume = {40}, number = {8}, pages = {865-878.e6}, year = {2022}, issn = {1535-6108}, doi = {https://doi.org/10.1016/j.ccell.2022.07.004}, url = {https://www.sciencedirect.com/science/article/pii/S1535610822003178}, author = {Chen, Richard J and Lu, Ming Y and Williamson, Drew FK and Chen, Tiffany Y and Lipkova, Jana and Shaban, Muhammad and Shady, Maha and Williams, Mane and Joo, Bumjin and Noor, Zahra and Mahmood, Faisal}, keywords = {deep learning, artificial intelligence, multimodal integration, cancer prognosis, multimodal prognostic models, pan-cancer, biomarker discovery, data fusion, computational pathology, digital pathology}, arxiv = {2108.02278}, code = {https://github.com/mahmoodlab/PORPOISE}, demo = {http://pancancer.mahmoodlab.org}, selected = {true}, abbr = {chen2022pan.png}, honor = {Best Paper, Case Western Artificial Intelligence in Oncology Symposium, 2020. Cover Art of Cancer Cell (Volume 40 Issue 8).} }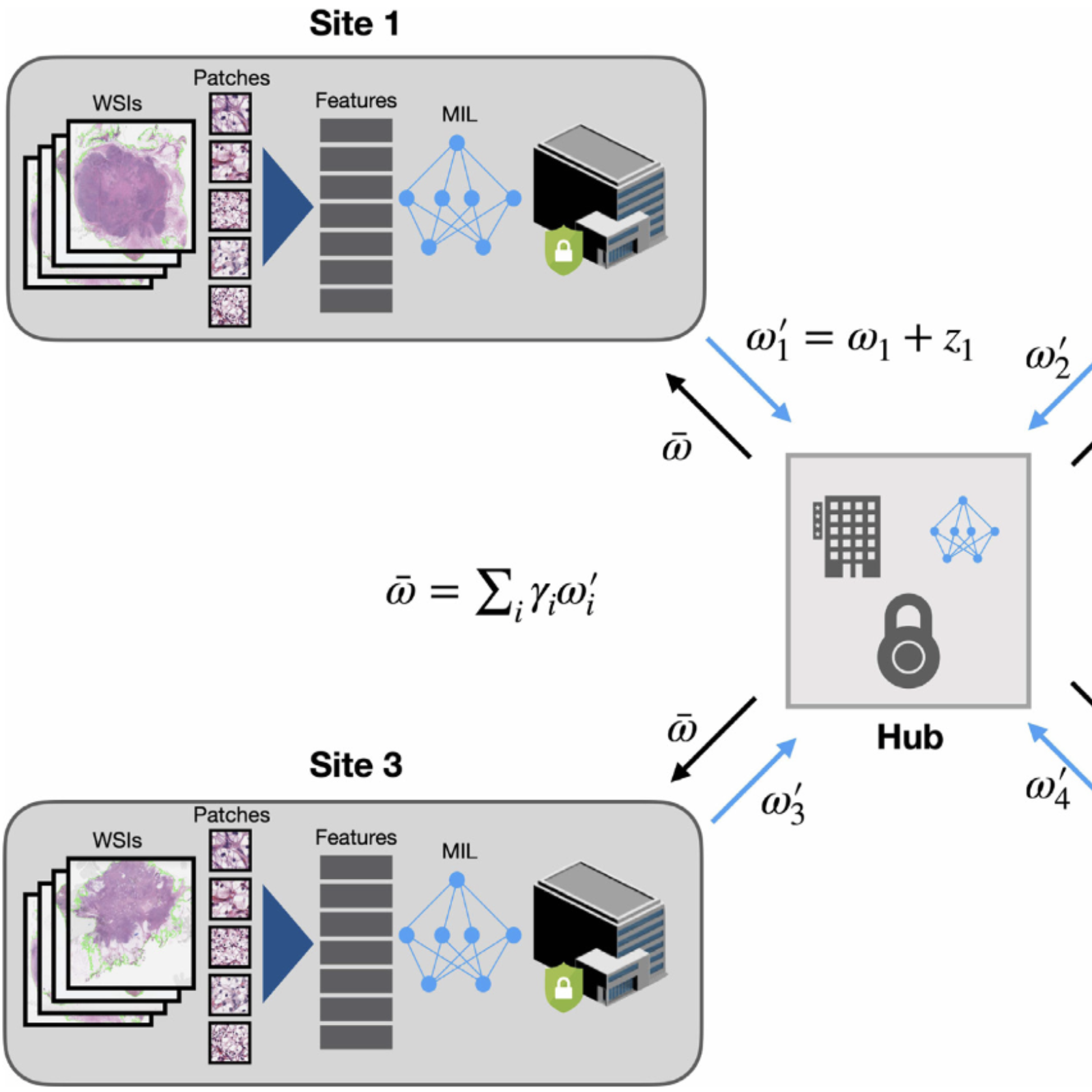 Federated Learning for Computational Pathology on Gigapixel Whole Slide ImagesMing Y. Lu*, Richard J. Chen*, Dehan Kong, Jana Lipkova, Rajendra Singh, Drew FK. Williamson, Tiffany Y. Chen, and Faisal MahmoodMedical Image Analysis 2022
Federated Learning for Computational Pathology on Gigapixel Whole Slide ImagesMing Y. Lu*, Richard J. Chen*, Dehan Kong, Jana Lipkova, Rajendra Singh, Drew FK. Williamson, Tiffany Y. Chen, and Faisal MahmoodMedical Image Analysis 2022Deep Learning-based computational pathology algorithms have demonstrated profound ability to excel in a wide array of tasks that range from characterization of well known morphological phenotypes to predicting non-human-identifiable features from histology such as molecular alterations. However, the development of robust, adaptable, and accurate deep learning-based models often rely on the collection and time-costly curation large high-quality annotated training data that should ideally come from diverse sources and patient populations to cater for the heterogeneity that exists in such datasets. Multi-centric and collaborative integration of medical data across multiple institutions can naturally help overcome this challenge and boost the model performance but is limited by privacy concerns amongst other difficulties that may arise in the complex data sharing process as models scale towards using hundreds of thousands of gigapixel whole slide images. In this paper, we introduce privacy-preserving federated learning for gigapixel whole slide images in computational pathology using weakly-supervised attention multiple instance learning and differential privacy. We evaluated our approach on two different diagnostic problems using thousands of histology whole slide images with only slide-level labels. Additionally, we present a weakly-supervised learning framework for survival prediction and patient stratification from whole slide images and demonstrate its effectiveness in a federated setting. Our results show that using federated learning, we can effectively develop accurate weakly supervised deep learning models from distributed data silos without direct data sharing and its associated complexities, while also preserving differential privacy using randomized noise generation.
@article{lu2022federated, title = {Federated Learning for Computational Pathology on Gigapixel Whole Slide Images}, url = {https://www.sciencedirect.com/science/article/pii/S1361841521003431}, author = {Lu*, Ming Y. and Chen*, Richard J. and Kong, Dehan and Lipkova, Jana and Singh, Rajendra and Williamson, Drew FK. and Chen, Tiffany Y. and Mahmood, Faisal}, journal = {Medical Image Analysis}, volume = {76}, pages = {102298}, year = {2022}, publisher = {Elsevier}, code = {https://github.com/mahmoodlab/HistoFL}, abbr = {lu2022histofl.png}, selected = {true}, arxiv = {2009.10190} }
2021
 Self-Supervised Vision Transformers Learn Visual Concepts in HistopathologyRichard J Chen, and Rahul G KrishnanLearning Meaningful Representations of Life, NeurIPS 2021
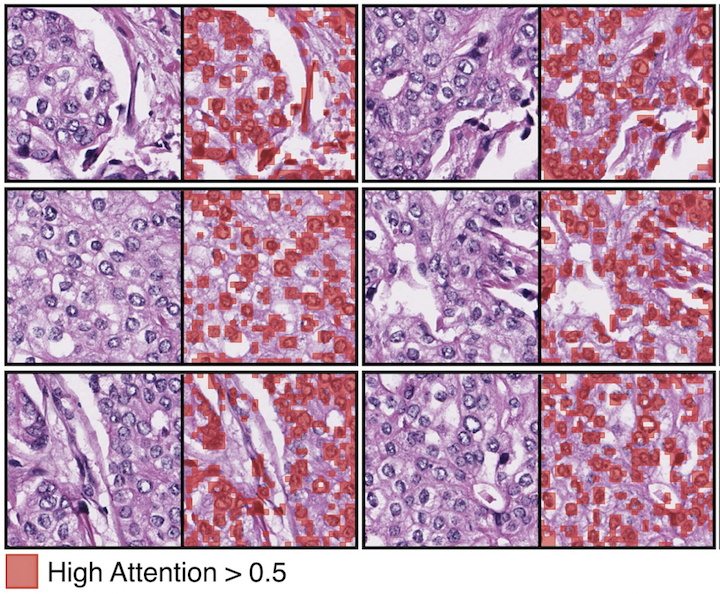
Self-Supervised Vision Transformers Learn Visual Concepts in HistopathologyRichard J Chen, and Rahul G KrishnanLearning Meaningful Representations of Life, NeurIPS 2021Tissue phenotyping is a fundamental task in learning objective characterizations of histopathologic biomarkers within the tumor-immune microenvironment in cancer pathology. However, whole-slide imaging (WSI) is a complex computer vision in which: 1) WSIs have enormous image resolutions with precludes large-scale pixel-level efforts in data curation, and 2) diversity of morphological phenotypes results in inter- and intra-observer variability in tissue labeling. To address these limitations, current efforts have proposed using pretrained image encoders (transfer learning from ImageNet, self-supervised pretraining) in extracting morphological features from pathology, but have not been extensively validated. In this work, we conduct a search for good representations in pathology by training a variety of self-supervised models with validation on a variety of weakly-supervised and patch-level tasks. Our key finding is in discovering that Vision Transformers using DINO-based knowledge distillation are able to learn data-efficient and interpretable features in histology images wherein the different attention heads learn distinct morphological phenotypes. We make evaluation code and pretrained weights publicly-available at: https://github.com/Richarizardd/Self-Supervised-ViT-Path.
@article{chen2022self, title = {Self-Supervised Vision Transformers Learn Visual Concepts in Histopathology}, author = {Chen, Richard J and Krishnan, Rahul G}, journal = {Learning Meaningful Representations of Life, NeurIPS}, year = {2021}, arxiv = {2203.00585}, code = {https://github.com/Richarizardd/Self-Supervised-ViT-Path}, abbr = {chen2022self.png} }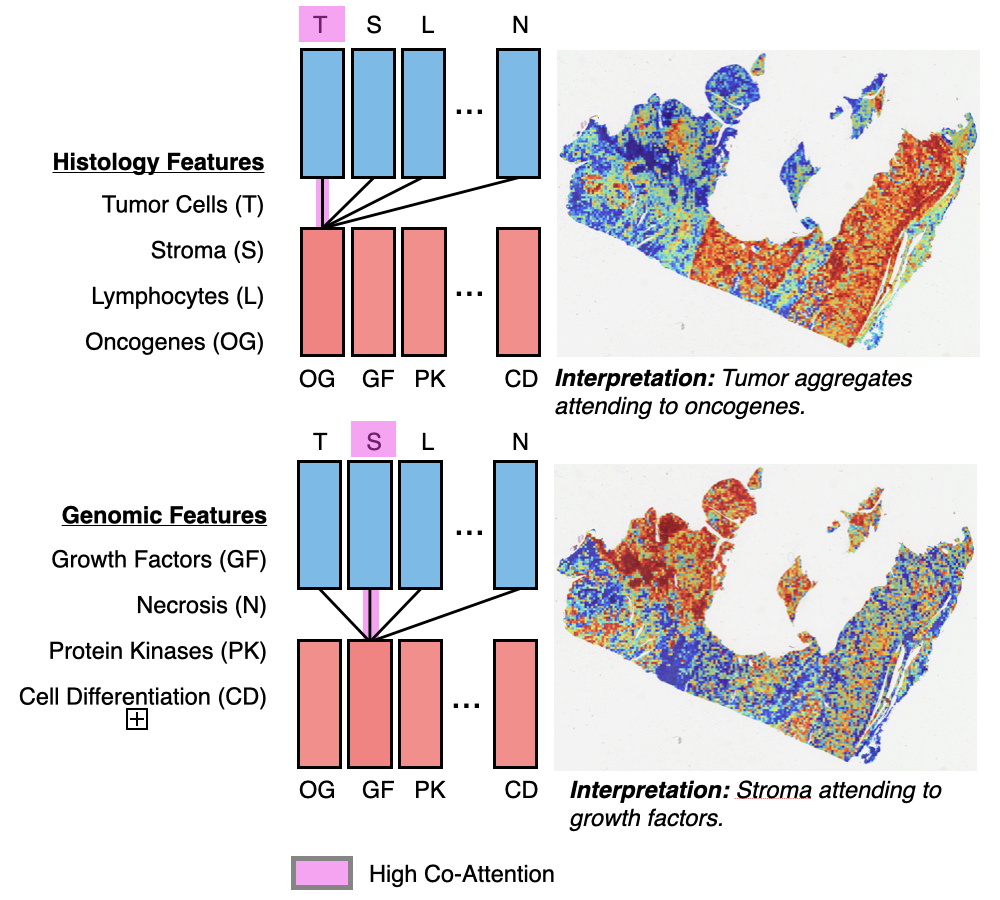 Multimodal Co-Attention Transformer for Survival Prediction in Gigapixel Whole Slide ImagesRichard J. Chen, Ming Y. Lu, Wei H. Weng, Tiffany Y Chen, Drew FK Williamson, Trevor Manz, Maha Shady, and Faisal MahmoodIn Proceedings of the IEEE/CVF International Conference on Computer Vision (ICCV) 2021
Multimodal Co-Attention Transformer for Survival Prediction in Gigapixel Whole Slide ImagesRichard J. Chen, Ming Y. Lu, Wei H. Weng, Tiffany Y Chen, Drew FK Williamson, Trevor Manz, Maha Shady, and Faisal MahmoodIn Proceedings of the IEEE/CVF International Conference on Computer Vision (ICCV) 2021Survival outcome prediction is a challenging weakly-supervised and ordinal regression task in computational pathology that involves modeling complex interactions within the tumor microenvironment in gigapixel whole slide images (WSIs). Despite recent progress in formulating WSIs as bags for multiple instance learning (MIL), representation learning of entire WSIs remains an open and challenging problem, especially in overcoming: 1) the computational complexity of feature aggregation in large bags, and 2) the data heterogeneity gap in incorporating biological priors such as genomic measurements. In this work, we present a Multimodal Co-Attention Transformer (MCAT) framework that learns an interpretable, dense co-attention mapping between WSIs and genomic features formulated in an embedding space. Inspired by approaches in Visual Question Answering (VQA) that can attribute how word embeddings attend to salient objects in an image when answering a question, MCAT learns how histology patches attend to genes when predicting patient survival. In addition to visualizing multimodal interactions, our co-attention transformation also reduces the space complexity of WSI bags, which enables the adaptation of Transformer layers as a general encoder backbone in MIL. We apply our proposed method on five different cancer datasets (4,730 WSIs, 67 million patches). Our experimental results demonstrate that the proposed method consistently achieves superior performance compared to the state-of-the-art methods.
@inproceedings{chen2021multimodal, title = {Multimodal Co-Attention Transformer for Survival Prediction in Gigapixel Whole Slide Images}, url = {https://openaccess.thecvf.com/content/ICCV2021/html/Chen_Multimodal_Co-Attention_Transformer_for_Survival_Prediction_in_Gigapixel_Whole_Slide_ICCV_2021_paper.html}, author = {Chen, Richard J. and Lu, Ming Y. and Weng, Wei H. and and Tiffany Y Chen and Williamson, Drew FK and Manz, Trevor and Shady, Maha and Mahmood, Faisal}, booktitle = {Proceedings of the IEEE/CVF International Conference on Computer Vision (ICCV)}, pages = {4015--4025}, year = {2021}, code = {https://github.com/mahmoodlab/MCAT}, abbr = {chen2021multimodal.png}, selected = {true} } Synthetic Data in Machine Learning for Medicine and HealthcareRichard J. Chen, Ming Y. Lu, Tiffany Y. Chen, Drew F. K. Williamson, and Faisal MahmoodNature Biomedical Engineering 2021
Synthetic Data in Machine Learning for Medicine and HealthcareRichard J. Chen, Ming Y. Lu, Tiffany Y. Chen, Drew F. K. Williamson, and Faisal MahmoodNature Biomedical Engineering 2021The proliferation of synthetic data in artificial intelligence for medicine and healthcare raises concerns about the vulnerabilities of the software and the challenges of current policy.
@article{chen2021synthetic, doi = {10.1038/s41551-021-00751-8}, url = {https://doi.org/10.1038/s41551-021-00751-8}, year = {2021}, month = jun, publisher = {Springer Science and Business Media {LLC}}, volume = {5}, number = {6}, pages = {493--497}, author = {Chen, Richard J. and Lu, Ming Y. and Chen, Tiffany Y. and Williamson, Drew F. K. and Mahmood, Faisal}, title = {Synthetic Data in Machine Learning for Medicine and Healthcare}, journal = {Nature Biomedical Engineering}, abbr = {chen2021synthetic.png}, selected = {true} }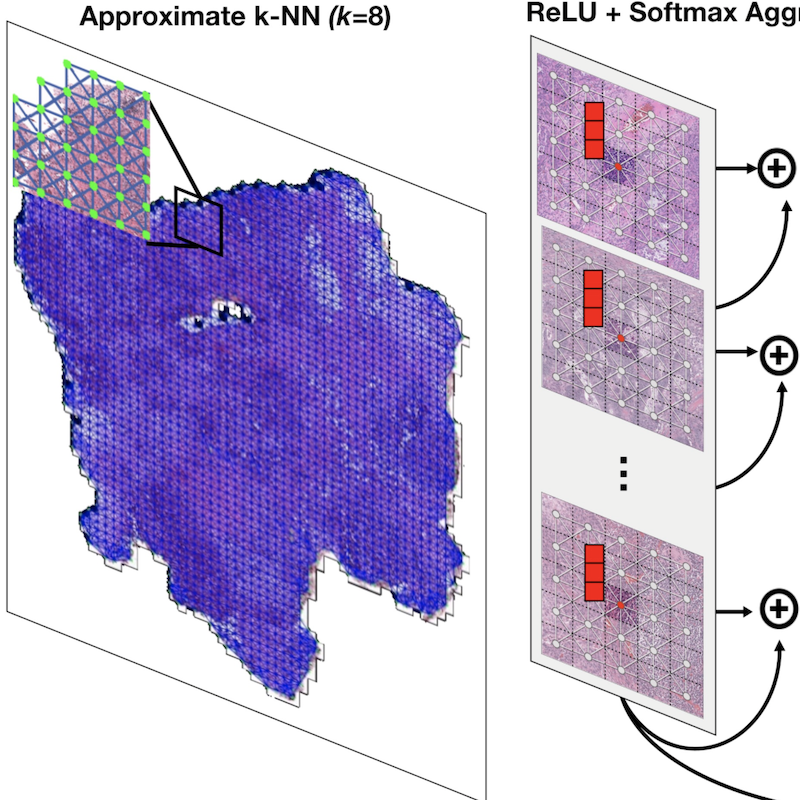 Whole Slide Images are 2D Point Clouds: Context-Aware Survival Prediction using Patch-Based Graph Convolutional NetworksRichard J. Chen, Ming Y. Lu, Muhammad Shaban, Chengkuan Chen, Tiffany Y Chen, Drew FK Williamson, and Faisal MahmoodIn International Conference on Medical Image Computing and Computer-Assisted Intervention (MICCAI) 2021
Whole Slide Images are 2D Point Clouds: Context-Aware Survival Prediction using Patch-Based Graph Convolutional NetworksRichard J. Chen, Ming Y. Lu, Muhammad Shaban, Chengkuan Chen, Tiffany Y Chen, Drew FK Williamson, and Faisal MahmoodIn International Conference on Medical Image Computing and Computer-Assisted Intervention (MICCAI) 2021Cancer prognostication is a challenging task in computational pathology that requires context-aware representations of histology features to adequately infer patient survival. Despite the advancements made in weakly-supervised deep learning, many approaches are not context-aware and are unable to model important morphological feature interactions between cell identities and tissue types that are prognostic for patient survival. In this work, we present Patch-GCN, a context-aware, spatially-resolved patch-based graph convolutional network that hierarchically aggregates instance-level histology features to model local- and global-level topological structures in the tumor microenvironment. We validate Patch-GCN with 4,370 gigapixel WSIs across five different cancer types from the Cancer Genome Atlas (TCGA), and demonstrate that Patch-GCN outperforms all prior weakly-supervised approaches by 3.58-9.46%. Our code and corresponding models are publicly available at this https URL: https://github.com/mahmoodlab/Patch-GCN.
@inproceedings{chen2021whole, title = {Whole Slide Images are 2D Point Clouds: Context-Aware Survival Prediction using Patch-Based Graph Convolutional Networks}, author = {Chen, Richard J. and Lu, Ming Y. and Shaban, Muhammad and Chen, Chengkuan and Chen, Tiffany Y and Williamson, Drew FK and Mahmood, Faisal}, booktitle = {International Conference on Medical Image Computing and Computer-Assisted Intervention (MICCAI)}, year = {2021}, organization = {Springer}, arxiv = {2107.13048}, abbr = {chen2021whole.png}, code = {https://github.com/mahmoodlab/Patch-GCN}, oral = {https://www.youtube.com/watch?v=KHv7ccs9FAI} }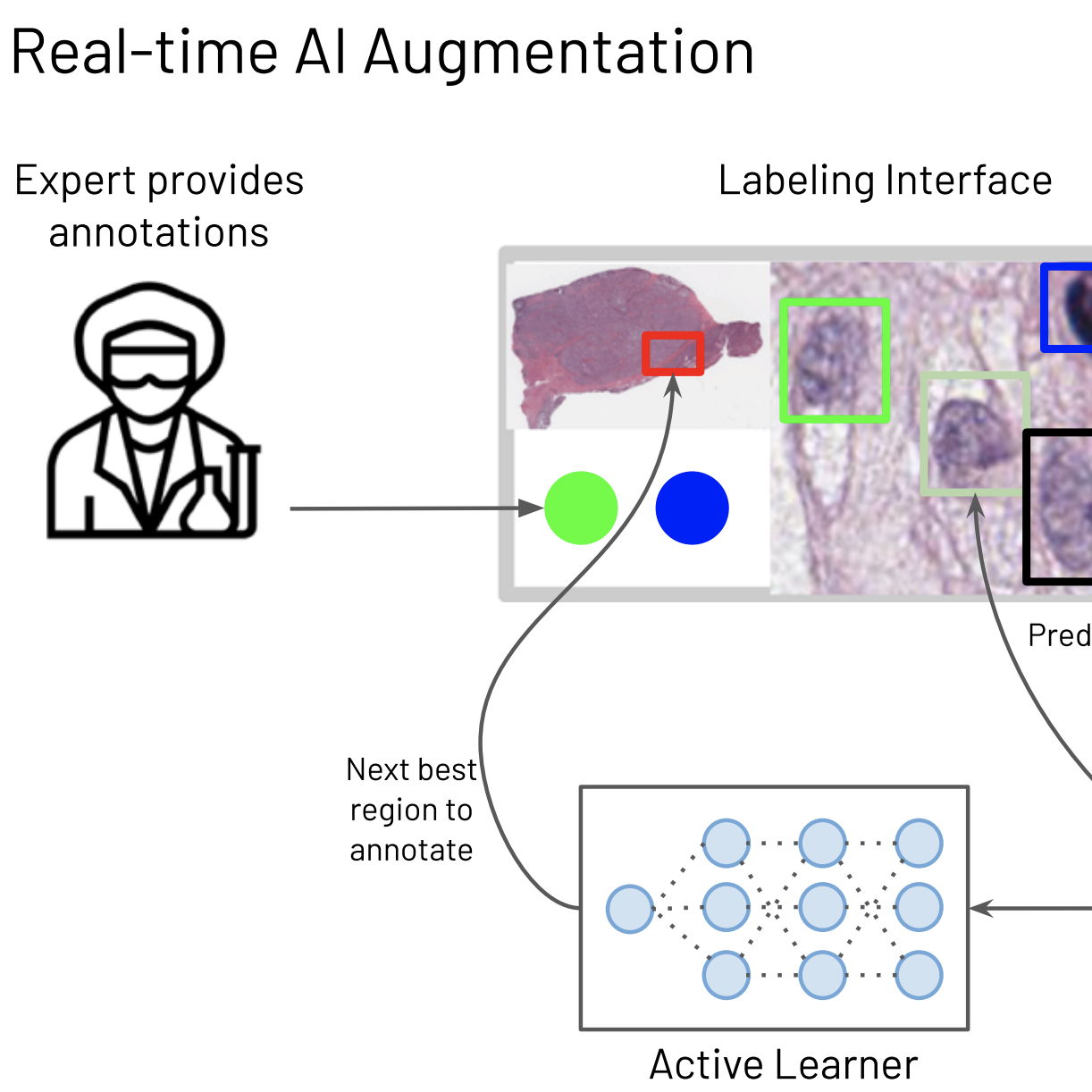 Efficient Cellular Annotation of Histopathology Slides with Real-Time AI augmentationJames A. Diao*, Richard J. Chen*, and Joseph C. Kvedarnpj Digital Medicine 2021
Efficient Cellular Annotation of Histopathology Slides with Real-Time AI augmentationJames A. Diao*, Richard J. Chen*, and Joseph C. Kvedarnpj Digital Medicine 2021In recent years, a steady swell of biological image data has driven rapid progress in healthcare applications of computer vision and machine learning. To make sense of this data, scientists often rely on detailed annotations from domain experts for training artificial intelligence (AI) algorithms. The time-consuming and costly process of collecting annotations presents a sizable bottleneck for AI research and development. HALS (Human-Augmenting Labeling System) is a collaborative human-AI labeling workflow that uses an iterative “review-and-revise” model to improve the efficiency of this critical process in computational pathology.
@article{diao2021efficient, doi = {10.1038/s41746-021-00534-0}, url = {https://doi.org/10.1038/s41746-021-00534-0}, year = {2021}, month = nov, publisher = {Springer Science and Business Media {LLC}}, volume = {4}, number = {1}, author = {Diao*, James A. and Chen*, Richard J. and Kvedar, Joseph C.}, title = {Efficient Cellular Annotation of Histopathology Slides with Real-Time {AI} augmentation}, journal = {npj Digital Medicine}, abbr = {diao2021efficient.png} }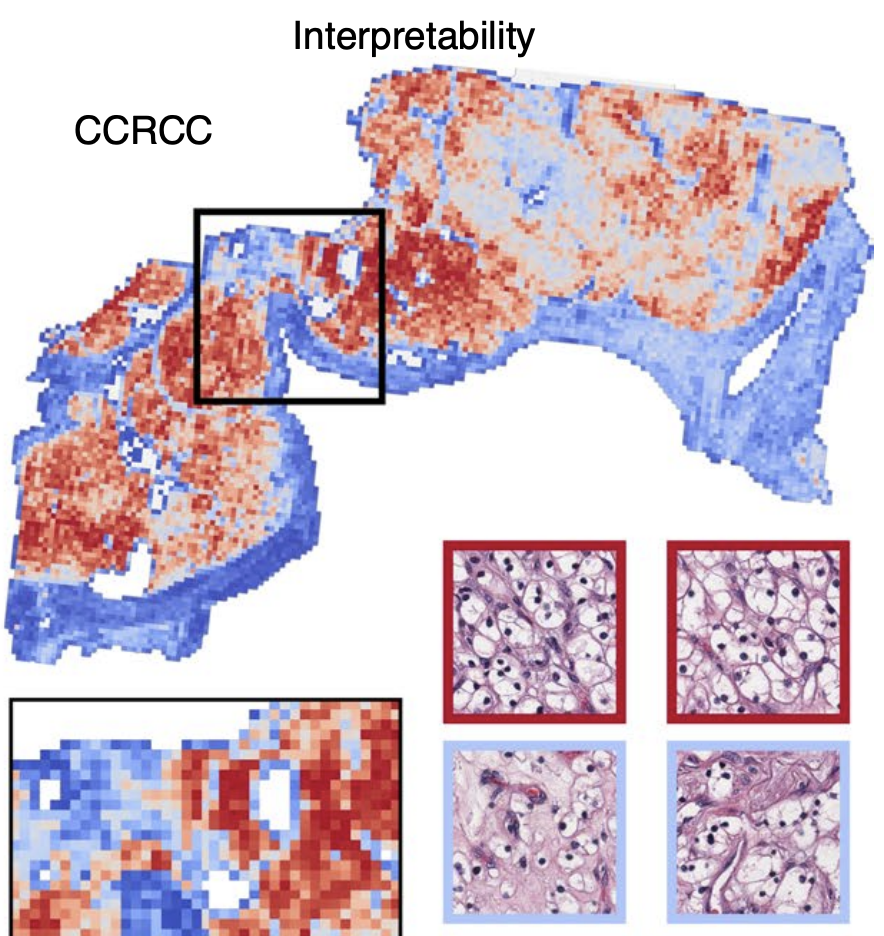 Data-Efficient and Weakly Supervised Computational Pathology on Whole-Slide ImagesMing Y. Lu, Drew FK. Williamson, Tiffany Y. Chen, Richard J. Chen, Matteo Barbieri, and Faisal MahmoodNature Biomedical Engineering 2021
Data-Efficient and Weakly Supervised Computational Pathology on Whole-Slide ImagesMing Y. Lu, Drew FK. Williamson, Tiffany Y. Chen, Richard J. Chen, Matteo Barbieri, and Faisal MahmoodNature Biomedical Engineering 2021Deep-learning methods for computational pathology require either manual annotation of gigapixel whole-slide images (WSIs) or large datasets of WSIs with slide-level labels and typically suffer from poor domain adaptation and interpretability. Here we report an interpretable weakly supervised deep-learning method for data-efficient WSI processing and learning that only requires slide-level labels. The method, which we named clustering-constrained-attention multiple-instance learning (CLAM), uses attention-based learning to identify subregions of high diagnostic value to accurately classify whole slides and instance-level clustering over the identified representative regions to constrain and refine the feature space. By applying CLAM to the subtyping of renal cell carcinoma and non-small-cell lung cancer as well as the detection of lymph node metastasis, we show that it can be used to localize well-known morphological features on WSIs without the need for spatial labels, that it overperforms standard weakly supervised classification algorithms and that it is adaptable to independent test cohorts, smartphone microscopy and varying tissue content.
@article{lu2021data, doi = {10.1038/s41551-020-00682-w}, url = {https://doi.org/10.1038/s41551-020-00682-w}, year = {2021}, month = mar, publisher = {Springer Science and Business Media {LLC}}, volume = {5}, number = {6}, pages = {555--570}, author = {Lu, Ming Y. and Williamson, Drew FK. and Chen, Tiffany Y. and Chen, Richard J. and Barbieri, Matteo and Mahmood, Faisal}, title = {Data-Efficient and Weakly Supervised Computational Pathology on Whole-Slide Images}, journal = {Nature Biomedical Engineering}, abbr = {lu2021data.png}, code = {https://github.com/mahmoodlab/CLAM}, arxiv = {2004.09666} }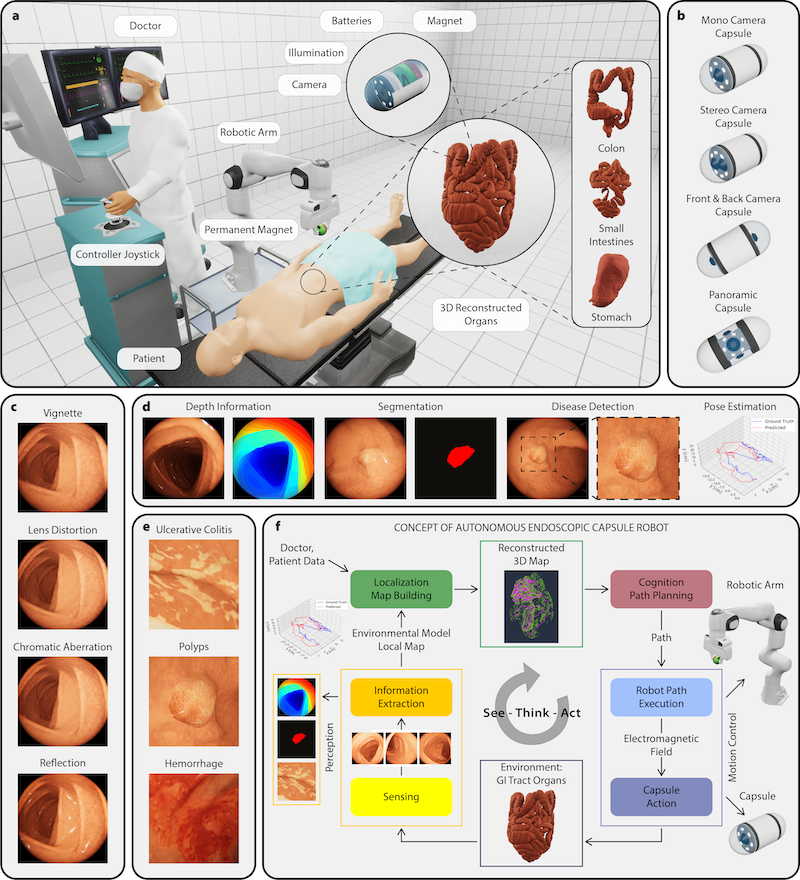 VR-Caps: A Virtual Environment for Capsule EndoscopyKağan İncetan, Ibrahim Omer Celik, Abdulhamid Obeid, Guliz Irem Gokceler, Kutsev Bengisu Ozyoruk, Yasin Almalioglu, Richard J Chen, Faisal Mahmood, Hunter Gilbert, Nicholas J Durr, and Mehmet TuranMedical Image Analysis 2021
VR-Caps: A Virtual Environment for Capsule EndoscopyKağan İncetan, Ibrahim Omer Celik, Abdulhamid Obeid, Guliz Irem Gokceler, Kutsev Bengisu Ozyoruk, Yasin Almalioglu, Richard J Chen, Faisal Mahmood, Hunter Gilbert, Nicholas J Durr, and Mehmet TuranMedical Image Analysis 2021Current capsule endoscopes and next-generation robotic capsules for diagnosis and treatment of gastrointestinal diseases are complex cyber-physical platforms that must orchestrate complex software and hardware functions. The desired tasks for these systems include visual localization, depth estimation, 3D mapping, disease detection and segmentation, automated navigation, active control, path realization and optional therapeutic modules such as targeted drug delivery and biopsy sampling. Data-driven algorithms promise to enable many advanced functionalities for capsule endoscopes, but real-world data is challenging to obtain. Physically-realistic simulations providing synthetic data have emerged as a solution to the development of data-driven algorithms. In this work, we present a comprehensive simulation platform for capsule endoscopy operations and introduce VR-Caps, a virtual active capsule environment that simulates a range of normal and abnormal tissue conditions (e.g., inflated, dry, wet etc.) and varied organ types, capsule endoscope designs (e.g., mono, stereo, dual and 360 camera), and the type, number, strength, and placement of internal and external magnetic sources that enable active locomotion. VR-Caps makes it possible to both independently or jointly develop, optimize, and test medical imaging and analysis software for the current and next-generation endoscopic capsule systems. To validate this approach, we train state-of-the-art deep neural networks to accomplish various medical image analysis tasks using simulated data from VR-Caps and evaluate the performance of these models on real medical data. Results demonstrate the usefulness and effectiveness of the proposed virtual platform in developing algorithms that quantify fractional coverage, camera trajectory, 3D map reconstruction, and disease classification. All of the code, pre-trained weights and created 3D organ models of the virtual environment with detailed instructions how to setup and use the environment are made publicly available at https://github.com/CapsuleEndoscope/VirtualCapsuleEndoscopy and a video demonstration can be seen in the supplementary videos (Video-I).
@article{incetan2021vr, doi = {10.1016/j.media.2021.101990}, url = {https://doi.org/10.1016/j.media.2021.101990}, year = {2021}, month = may, publisher = {Elsevier {BV}}, volume = {70}, pages = {101990}, title = {VR-Caps: A Virtual Environment for Capsule Endoscopy}, author = {{\.I}ncetan, Ka{\u{g}}an and Celik, Ibrahim Omer and Obeid, Abdulhamid and Gokceler, Guliz Irem and Ozyoruk, Kutsev Bengisu and Almalioglu, Yasin and Chen, Richard J and Mahmood, Faisal and Gilbert, Hunter and Durr, Nicholas J and Turan, Mehmet}, journal = {Medical Image Analysis}, code = {https://github.com/CapsuleEndoscope/VirtualCapsuleEndoscopy}, arxiv = {2008.12949}, abbr = {incetan2021vr.png}, demo = {https://www.youtube.com/watch?v=UQ2u3CIUciA} }
2020
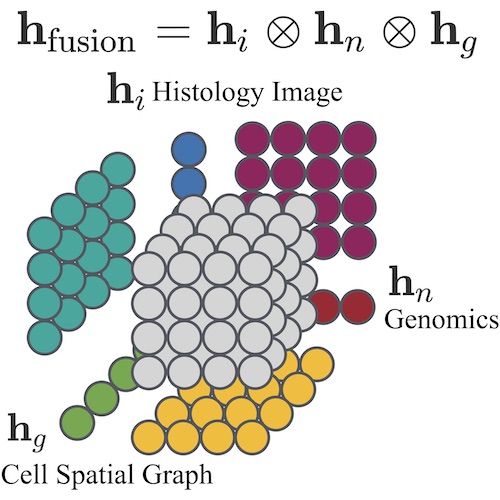 Pathomic Fusion: An Integrated Framework for Fusing Histopathology and Genomic Features for Cancer Diagnosis and PrognosisRichard J. Chen, Ming Y. Lu, Jingwen Wang, Drew F. K. Williamson, Scott J. Rodig, Neal I. Lindeman, and Faisal MahmoodIEEE Transactions on Medical Imaging 2020Top 5 Posters, NVIDIA GTC 2020
Pathomic Fusion: An Integrated Framework for Fusing Histopathology and Genomic Features for Cancer Diagnosis and PrognosisRichard J. Chen, Ming Y. Lu, Jingwen Wang, Drew F. K. Williamson, Scott J. Rodig, Neal I. Lindeman, and Faisal MahmoodIEEE Transactions on Medical Imaging 2020Top 5 Posters, NVIDIA GTC 2020Cancer diagnosis, prognosis, and therapeutic response predictions are based on morphological information from histology slides and molecular profiles from genomic data. However, most deep learning-based objective outcome prediction and grading paradigms are based on histology or genomics alone and do not make use of the complementary information in an intuitive manner. In this work, we propose Pathomic Fusion, an interpretable strategy for end-to-end multimodal fusion of histology image and genomic (mutations, CNV, RNASeq) features for survival outcome prediction. Our approach models pairwise feature interactions across modalities by taking the Kronecker product of unimodal feature representations, and controls the expressiveness of each representation via a gatingbased attention mechanism. Following supervised learning, we are able to interpret and saliently localize features across each modality, and understand how feature importance shifts when conditioning on multimodal input. We validate our approach using glioma and clear cell renal cell carcinoma datasets from the Cancer Genome Atlas (TCGA), which contains paired wholeslide image, genotype, and transcriptome data with ground truth survival and histologic grade labels. In a 15-fold cross-validation, our results demonstrate that the proposed multimodal fusion paradigm improves prognostic determinations from ground truth grading and molecular subtyping, as well as unimodal deep networks trained on histology and genomic data alone. The proposed method establishes insight and theory on how to train deep networks on multimodal biomedical data in an intuitive manner, which will be useful for other problems in medicine that seek to combine heterogeneous data streams for understanding diseases and predicting response and resistance to treatment. Code and trained models are made available at: https://github.com/mahmoodlab/PathomicFusion.
@article{chen2019pathomic, doi = {10.1109/tmi.2020.3021387}, year = {2020}, publisher = {Institute of Electrical and Electronics Engineers ({IEEE})}, pages = {1--1}, author = {Chen, Richard J. and Lu, Ming Y. and Wang, Jingwen and Williamson, Drew F. K. and Rodig, Scott J. and Lindeman, Neal I. and Mahmood, Faisal}, title = {Pathomic Fusion: An Integrated Framework for Fusing Histopathology and Genomic Features for Cancer Diagnosis and Prognosis}, journal = {{IEEE} Transactions on Medical Imaging}, arxiv = {1912.08937}, abbr = {chen2019pathomic.png}, honor = {Top 5 Posters, NVIDIA GTC 2020}, oral = {https://www.youtube.com/watch?v=TrjGEUVX5YE}, code = {https://github.com/mahmoodlab/PathomicFusion}, press = {https://blogs.nvidia.com/blog/2019/11/07/harvard-pathology-lab-data-fusion-ai-cancer/}, selected = {true} }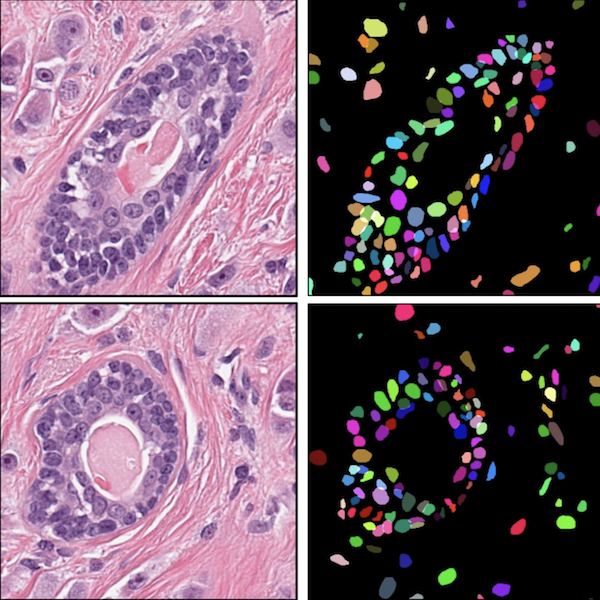 Deep Adversarial Training for Multi-Organ Nuclei Segmentation in Histopathology ImagesFaisal Mahmood, Daniel Borders, Richard J. Chen, Gregory N. Mckay, Kevan J. Salimian, Alexander Baras, and Nicholas J. DurrIEEE Transactions on Medical Imaging 2020
Deep Adversarial Training for Multi-Organ Nuclei Segmentation in Histopathology ImagesFaisal Mahmood, Daniel Borders, Richard J. Chen, Gregory N. Mckay, Kevan J. Salimian, Alexander Baras, and Nicholas J. DurrIEEE Transactions on Medical Imaging 2020Nuclei mymargin segmentation is a fundamental task for various computational pathology applications including nuclei morphology analysis, cell type classification, and cancer grading. Deep learning has emerged as a powerful approach to segmenting nuclei but the accuracy of convolutional neural networks (CNNs) depends on the volume and the quality of labeled histopathology data for training. In particular, conventional CNN-based approaches lack structured prediction capabilities, which are required to distinguish overlapping and clumped nuclei. Here, we present an approach to nuclei segmentation that overcomes these challenges by utilizing a conditional generative adversarial network (cGAN) trained with synthetic and real data. We generate a large dataset of H&E training images with perfect nuclei segmentation labels using an unpaired GAN framework. This synthetic data along with real histopathology data from six different organs are used to train a conditional GAN with spectral normalization and gradient penalty for nuclei segmentation. This adversarial regression framework enforces higher-order spacial-consistency when compared to conventional CNN models. We demonstrate that this nuclei segmentation approach generalizes across different organs, sites, patients and disease states, and outperforms conventional approaches, especially in isolating individual and overlapping nuclei.
@article{mahmood2018seg, doi = {10.1109/tmi.2019.2927182}, url = {https://doi.org/10.1109/tmi.2019.2927182}, year = {2020}, month = nov, publisher = {Institute of Electrical and Electronics Engineers ({IEEE})}, volume = {39}, number = {11}, pages = {3257--3267}, author = {Mahmood, Faisal and Borders, Daniel and Chen, Richard J. and Mckay, Gregory N. and Salimian, Kevan J. and Baras, Alexander and Durr, Nicholas J.}, title = {Deep Adversarial Training for Multi-Organ Nuclei Segmentation in Histopathology Images}, journal = {{IEEE} Transactions on Medical Imaging}, abbr = {mahmood2018seg.png}, code = {https://github.com/mahmoodlab/NucleiSegmentation}, arxiv = {1810.00236} }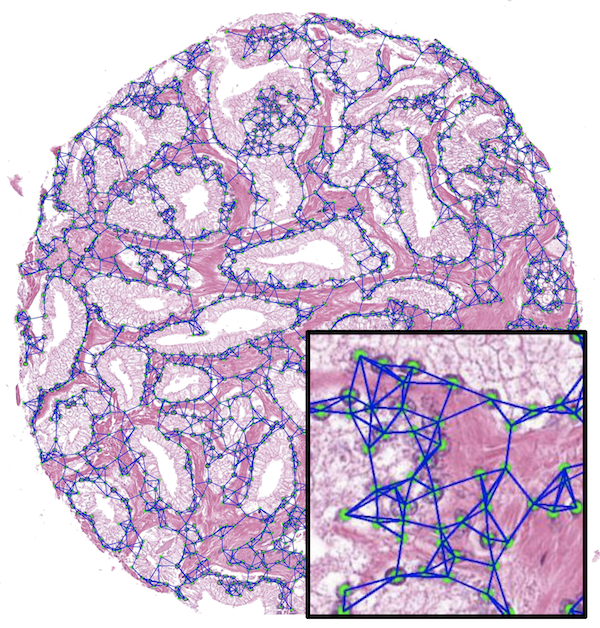 Weakly Supervised Prostate TMA Classification via Graph Convolutional NetworksJingwen Wang, Richard J Chen, Ming Y Lu, Alexander Baras, and Faisal MahmoodIn 2020 IEEE 17th International Symposium on Biomedical Imaging (ISBI) 2020Oral Presentation
Weakly Supervised Prostate TMA Classification via Graph Convolutional NetworksJingwen Wang, Richard J Chen, Ming Y Lu, Alexander Baras, and Faisal MahmoodIn 2020 IEEE 17th International Symposium on Biomedical Imaging (ISBI) 2020Oral PresentationHistology-based grade classification is clinically important for many cancer types in stratifying patients distinct treatment groups. In prostate cancer, the Gleason score is a grading system used to measure the aggressiveness of prostate cancer from the spatial organization of cells and the distribution of glands. However, the subjective interpretation of Gleason score often suffers from large interobserver and intraobserver variability. Previous work in deep learning-based objective Gleason grading requires manual pixel-level annotation. In this work, we propose a weakly-supervised approach for grade classification in tissue micro-arrays (TMA) using graph convolutional networks (GCNs), in which we model the spatial organization of cells as a graph to better capture the proliferation and community structure of tumor cells. As node-level features in our graph representation, we learn the morphometry of each cell using a contrastive predictive coding (CPC)-based self-supervised approach. We demonstrate that on a five-fold cross validation our method can achieve 0.9659±0.0096 AUC using only TMA-level labels. Our method demonstrates a 39.80% improvement over standard GCNs with texture features and a 29.27% improvement over GCNs with VGG19 features. Our proposed pipeline can be used to objectively stratify low and high risk cases, reducing inter- and intra-observer variability and pathologist workload.
@inproceedings{wang2019weakly, doi = {10.1109/isbi45749.2020.9098534}, url = {https://doi.org/10.1109/isbi45749.2020.9098534}, year = {2020}, month = apr, publisher = {{IEEE}}, title = {Weakly Supervised Prostate TMA Classification via Graph Convolutional Networks}, author = {Wang, Jingwen and Chen, Richard J and Lu, Ming Y and Baras, Alexander and Mahmood, Faisal}, booktitle = {2020 {IEEE} 17th International Symposium on Biomedical Imaging ({ISBI})}, arxiv = {1910.13328}, honor = {Oral Presentation}, abbr = {wang2019weakly.png} }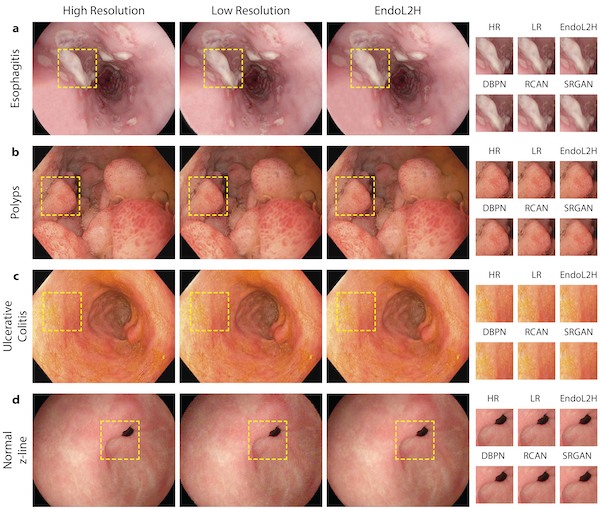 EndoL2H: Deep Super-Resolution for Capsule EndoscopyYasin Almalioglu, Kutsev Bengisu Ozyoruk, Abdulkadir Gokce, Kagan Incetan, Guliz Irem Gokceler, Muhammed Ali Simsek, Kivanc Ararat, Richard J Chen, Nicholas J Durr, Faisal Mahmood, and othersIEEE Transactions on Medical Imaging 2020
EndoL2H: Deep Super-Resolution for Capsule EndoscopyYasin Almalioglu, Kutsev Bengisu Ozyoruk, Abdulkadir Gokce, Kagan Incetan, Guliz Irem Gokceler, Muhammed Ali Simsek, Kivanc Ararat, Richard J Chen, Nicholas J Durr, Faisal Mahmood, and othersIEEE Transactions on Medical Imaging 2020Although wireless capsule endoscopy is the preferred modality for diagnosis and assessment of small bowel diseases, the poor camera resolution is a substantial limitation for both subjective and automated diagnostics. Enhanced-resolution endoscopy has shown to improve adenoma detection rate for conventional endoscopy and is likely to do the same for capsule endoscopy. In this work, we propose and quantitatively validate a novel framework to learn a mapping from low-to-high resolution endoscopic images. We combine conditional adversarial networks with a spatial attention block to improve the resolution by up to factors of 8x, 10x, 12x, respectively. Quantitative and qualitative studies performed demonstrate the superiority of EndoL2H over state-of-the-art deep super-resolution methods DBPN, RCAN and SRGAN. MOS tests performed by 30 gastroenterologists qualitatively assess and confirm the clinical relevance of the approach. EndoL2H is generally applicable to any endoscopic capsule system and has the potential to improve diagnosis and better harness computational approaches for polyp detection and characterization. Our code and trained models are available at https://github.com/CapsuleEndoscope/EndoL2H.
@article{almalioglu2020endol2h, doi = {10.1109/tmi.2020.3016744}, url = {https://doi.org/10.1109/tmi.2020.3016744}, year = {2020}, month = dec, publisher = {Institute of Electrical and Electronics Engineers ({IEEE})}, volume = {39}, number = {12}, pages = {4297--4309}, title = {{EndoL}2H: Deep Super-Resolution for Capsule Endoscopy}, journal = {{IEEE} Transactions on Medical Imaging}, author = {Almalioglu, Yasin and Ozyoruk, Kutsev Bengisu and Gokce, Abdulkadir and Incetan, Kagan and Gokceler, Guliz Irem and Simsek, Muhammed Ali and Ararat, Kivanc and Chen, Richard J and Durr, Nicholas J and Mahmood, Faisal and others}, arxiv = {2002.05459}, code = {https://github.com/CapsuleEndoscope/EndoL2H}, abbr = {almalioglu2020endol2h.png} }
2019
 Developing Measures of Cognitive Impairment in the Real World from Consumer-Grade Multimodal Sensor StreamsRichard J. Chen*, Filip Jankovic*, Nikki Marinsek*, Luca Foschini, Lampros Kourtis, Alessio Signorini, Melissa Pugh, Jie Shen, Roy Yaari, Vera Maljkovic, Marc Sunga, Han Hee Song, Hyun Joon Jung, Belle Tseng, and Andrew TristerIn Proceedings of the 25th ACM SIGKDD International Conference on Knowledge Discovery & Data Mining 2019Oral Presentation & Best Paper Runner-Up
Developing Measures of Cognitive Impairment in the Real World from Consumer-Grade Multimodal Sensor StreamsRichard J. Chen*, Filip Jankovic*, Nikki Marinsek*, Luca Foschini, Lampros Kourtis, Alessio Signorini, Melissa Pugh, Jie Shen, Roy Yaari, Vera Maljkovic, Marc Sunga, Han Hee Song, Hyun Joon Jung, Belle Tseng, and Andrew TristerIn Proceedings of the 25th ACM SIGKDD International Conference on Knowledge Discovery & Data Mining 2019Oral Presentation & Best Paper Runner-UpThe ubiquity and remarkable technological progress of wearable consumer devices and mobile-computing platforms (smart phone, smart watch, tablet), along with the multitude of sensor modalities available, have enabled continuous monitoring of patients and their daily activities. Such rich, longitudinal information can be mined for physiological and behavioral signatures of cognitive impairment and provide new avenues for detecting MCI in a timely and cost-effective manner. In this work, we present a platform for remote and unobtrusive monitoring of symptoms related to cognitive impairment using several consumer-grade smart devices. We demonstrate how the platform has been used to collect a total of 16TB of data during the Lilly Exploratory Digital Assessment Study, a 12-week feasibility study which monitored 31 people with cognitive impairment and 82 without cognitive impairment in free living conditions. We describe how careful data unification, time-alignment, and imputation techniques can handle missing data rates inherent in real-world settings and ultimately show utility of these disparate data in differentiating symptomatics from healthy controls based on features computed purely from device data.
@inproceedings{chen2019developing, doi = {10.1145/3292500.3330690}, url = {https://machinelearning.apple.com/research/developing-measures-of-cognitive-impairment-in-the-real-world-from-consumer-grade-multimodal-sensor-streams}, year = {2019}, month = jul, publisher = {{ACM}}, author = {Chen*, Richard J. and Jankovic*, Filip and Marinsek*, Nikki and Foschini, Luca and Kourtis, Lampros and Signorini, Alessio and Pugh, Melissa and Shen, Jie and Yaari, Roy and Maljkovic, Vera and Sunga, Marc and Song, Han Hee and Jung, Hyun Joon and Tseng, Belle and Trister, Andrew}, title = {Developing Measures of Cognitive Impairment in the Real World from Consumer-Grade Multimodal Sensor Streams}, booktitle = {Proceedings of the 25th {ACM} {SIGKDD} International Conference on Knowledge Discovery {\&} Data Mining}, honor = {Oral Presentation & Best Paper Runner-Up}, press = {https://www.technologyreview.com/2019/08/08/102821/your-apple-watch-might-one-day-spot-if-youre-developing-alzheimers/}, abbr = {chen2019developing.png}, oral = {https://www.youtube.com/watch?v=H_wTI4LUW7A}, selected = {true} } From Detection of Individual Metastases to Classification of Lymph Node Status at the Patient Level: The CAMELYON17 ChallengePeter Bandi, Oscar Geessink, Quirine Manson, Marcory Van Dijk, Maschenka Balkenhol, Meyke Hermsen, Babak Ehteshami Bejnordi, Byungjae Lee, Kyunghyun Paeng, Aoxiao Zhong, Quanzheng Li, Farhad Ghazvinian Zanjani, Svitlana Zinger, Keisuke Fukuta, Daisuke Komura, Vlado Ovtcharov, Shenghua Cheng, Shaoqun Zeng, Jeppe Thagaard, Anders B. Dahl, Huangjing Lin, Hao Chen, Ludwig Jacobsson, Martin Hedlund, Melih cetin, Eren Halici, Hunter Jackson, Richard J. Chen, Fabian Both, Jorg Franke, Heidi Kusters-Vandevelde, Willem Vreuls, Peter Bult, Bram Ginneken, Jeroen Laak, and Geert LitjensIEEE Transactions on Medical Imaging 20198th Place, CAMELYON17 Challenge
From Detection of Individual Metastases to Classification of Lymph Node Status at the Patient Level: The CAMELYON17 ChallengePeter Bandi, Oscar Geessink, Quirine Manson, Marcory Van Dijk, Maschenka Balkenhol, Meyke Hermsen, Babak Ehteshami Bejnordi, Byungjae Lee, Kyunghyun Paeng, Aoxiao Zhong, Quanzheng Li, Farhad Ghazvinian Zanjani, Svitlana Zinger, Keisuke Fukuta, Daisuke Komura, Vlado Ovtcharov, Shenghua Cheng, Shaoqun Zeng, Jeppe Thagaard, Anders B. Dahl, Huangjing Lin, Hao Chen, Ludwig Jacobsson, Martin Hedlund, Melih cetin, Eren Halici, Hunter Jackson, Richard J. Chen, Fabian Both, Jorg Franke, Heidi Kusters-Vandevelde, Willem Vreuls, Peter Bult, Bram Ginneken, Jeroen Laak, and Geert LitjensIEEE Transactions on Medical Imaging 20198th Place, CAMELYON17 ChallengeAutomated detection of cancer metastases in lymph nodes has the potential to improve the assessment of prognosis for patients. To enable fair comparison between the algorithms for this purpose, we set up the CAMELYON17 challenge in conjunction with the IEEE International Symposium on Biomedical Imaging 2017 Conference in Melbourne. Over 300 participants registered on the challenge website, of which 23 teams submitted a total of 37 algorithms before the initial deadline. Participants were provided with 899 whole-slide images (WSIs) for developing their algorithms. The developed algorithms were evaluated based on the test set encompassing 100 patients and 500 WSIs. The evaluation metric used was a quadratic weighted Cohen’s kappa. We discuss the algorithmic details of the 10 best pre-conference and two post-conference submissions. All these participants used convolutional neural networks in combination with pre- and postprocessing steps. Algorithms differed mostly in neural network architecture, training strategy, and pre- and postprocessing methodology. Overall, the kappa metric ranged from 0.89 to -0.13 across all submissions. The best results were obtained with pre-trained architectures such as ResNet. Confusion matrix analysis revealed that all participants struggled with reliably identifying isolated tumor cells, the smallest type of metastasis, with detection rates below 40%. Qualitative inspection of the results of the top participants showed categories of false positives, such as nerves or contamination, which could be targeted for further optimization. Last, we show that simple combinations of the top algorithms result in higher kappa metric values than any algorithm individually, with 0.93 for the best combination.
@article{bandi2019detection, doi = {10.1109/tmi.2018.2867350}, url = {https://doi.org/10.1109/tmi.2018.2867350}, year = {2019}, month = feb, publisher = {Institute of Electrical and Electronics Engineers ({IEEE})}, volume = {38}, number = {2}, pages = {550--560}, author = {Bandi, Peter and Geessink, Oscar and Manson, Quirine and Dijk, Marcory Van and Balkenhol, Maschenka and Hermsen, Meyke and Bejnordi, Babak Ehteshami and Lee, Byungjae and Paeng, Kyunghyun and Zhong, Aoxiao and Li, Quanzheng and Zanjani, Farhad Ghazvinian and Zinger, Svitlana and Fukuta, Keisuke and Komura, Daisuke and Ovtcharov, Vlado and Cheng, Shenghua and Zeng, Shaoqun and Thagaard, Jeppe and Dahl, Anders B. and Lin, Huangjing and Chen, Hao and Jacobsson, Ludwig and Hedlund, Martin and cetin, Melih and Halici, Eren and Jackson, Hunter and Chen, Richard J. and Both, Fabian and Franke, Jorg and Kusters-Vandevelde, Heidi and Vreuls, Willem and Bult, Peter and van Ginneken, Bram and van der Laak, Jeroen and Litjens, Geert}, title = {From Detection of Individual Metastases to Classification of Lymph Node Status at the Patient Level: The {CAMELYON}17 Challenge}, journal = {{IEEE} Transactions on Medical Imaging}, honor = {8th Place, CAMELYON17 Challenge}, demo = {https://www.youtube.com/watch?v=cfyhLMZl6qU}, abbr = {bandi2019detection.png} } SLAM Endoscopy Enhanced by Adversarial Depth PredictionRichard J. Chen, Taylor L Bobrow, Thomas Athey, Faisal Mahmood, and Nicholas J DurrIn DSHealth Workshop in the 25th ACM SIGKDD International Conference on Knowledge Discovery & Data Mining 2019Spotlight Presentation
SLAM Endoscopy Enhanced by Adversarial Depth PredictionRichard J. Chen, Taylor L Bobrow, Thomas Athey, Faisal Mahmood, and Nicholas J DurrIn DSHealth Workshop in the 25th ACM SIGKDD International Conference on Knowledge Discovery & Data Mining 2019Spotlight PresentationMedical endoscopy remains a challenging application for simultaneous localization and mapping (SLAM) due to the sparsity of image features and size constraints that prevent direct depth-sensing. We present a SLAM approach that incorporates depth predictions made by an adversarially-trained convolutional neural network (CNN) applied to monocular endoscopy images. The depth network is trained with synthetic images of a simple colon model, and then fine-tuned with domain-randomized, photorealistic images rendered from computed tomography measurements of human colons. Each image is paired with an error-free depth map for supervised adversarial learning. Monocular RGB images are then fused with corresponding depth predictions, enabling dense reconstruction and mosaicing as an endoscope is advanced through the gastrointestinal tract. Our preliminary results demonstrate that incorporating monocular depth estimation into a SLAM architecture can enable dense reconstruction of endoscopic scenes.
@inproceedings{chen2019slam, title = {SLAM Endoscopy Enhanced by Adversarial Depth Prediction}, author = {Chen, Richard J. and Bobrow, Taylor L and Athey, Thomas and Mahmood, Faisal and Durr, Nicholas J}, booktitle = {DSHealth Workshop in the 25th {ACM} {SIGKDD} International Conference on Knowledge Discovery {\&} Data Mining}, year = {2019}, abbr = {chen2019slam.png}, arxiv = {1907.00283}, honor = {Spotlight Presentation}, url = {https://dshealthkdd.github.io/dshealth-2019/#papers}, demo = {https://www.youtube.com/watch?v=7I-d5LwIAQI} } Semi-Supervised Histology Classification using Deep Multiple Instance Larning and Contrastive Predictive CodingMing Y Lu, Richard J Chen, Jingwen Wang, Debora Dillon, and Faisal MahmoodIn ML4H Workshop in Advances in Neural Information Processing Systems (NeurIPS) 2019Oral Prsentation in SPIE Medical Imaging 2020
Semi-Supervised Histology Classification using Deep Multiple Instance Larning and Contrastive Predictive CodingMing Y Lu, Richard J Chen, Jingwen Wang, Debora Dillon, and Faisal MahmoodIn ML4H Workshop in Advances in Neural Information Processing Systems (NeurIPS) 2019Oral Prsentation in SPIE Medical Imaging 2020Convolutional neural networks can be trained to perform histology slide classification using weak annotations with multiple instance learning (MIL). However, given the paucity of labeled histology data, direct application of MIL can easily suffer from overfitting and the network is unable to learn rich feature representations due to the weak supervisory signal. We propose to overcome such limitations with a two-stage semi-supervised approach that combines the power of data-efficient self-supervised feature learning via contrastive predictive coding (CPC) and the interpretability and flexibility of regularized attention-based MIL. We apply our two-stage CPC + MIL semi-supervised pipeline to the binary classification of breast cancer histology images. Across five random splits, we report state-of-the-art performance with a mean validation accuracy of 95% and an area under the ROC curve of 0.968. We further evaluate the quality of features learned via CPC relative to simple transfer learning and show that strong classification performance using CPC features can be efficiently leveraged under the MIL framework even with the feature encoder frozen.
@inproceedings{lu2019semi, title = {Semi-Supervised Histology Classification using Deep Multiple Instance Larning and Contrastive Predictive Coding}, author = {Lu, Ming Y and Chen, Richard J and Wang, Jingwen and Dillon, Debora and Mahmood, Faisal}, booktitle = {ML4H Workshop in Advances in Neural Information Processing Systems (NeurIPS)}, year = {2019}, arxiv = {1910.10825}, abbr = {lu2019semi.png}, doi = {10.1117/12.2549627}, url = {https://doi.org/10.1117/12.2549627}, oral = {https://www.spiedigitallibrary.org/conference-proceedings-of-spie/11320/113200J/Semi-supervised-breast-cancer-histology-classification-using-deep-multiple-instance/10.1117/12.2549627.short?SSO=1}, honor = {Oral Prsentation in SPIE Medical Imaging 2020} }
2018
 Precision of Regional Wall Motion Estimates from Ultra-Low-Dose Cardiac CT using SQUEEZAmir Pourmorteza, Noemie Keller, Richard J. Chen, Albert Lardo, Henry Halperin, Marcus Y. Chen, and Elliot McVeighThe International Journal of Cardiovascular Imaging 2018
Precision of Regional Wall Motion Estimates from Ultra-Low-Dose Cardiac CT using SQUEEZAmir Pourmorteza, Noemie Keller, Richard J. Chen, Albert Lardo, Henry Halperin, Marcus Y. Chen, and Elliot McVeighThe International Journal of Cardiovascular Imaging 2018Resting regional wall motion abnormality (RWMA) has significant prognostic value beyond the findings of computed tomography (CT) coronary angiography. Stretch quantification of endocardial engraved zones (SQUEEZ) has been proposed as a measure of regional cardiac function. The purpose of the work reported here was to determine the effect of lowering the radiation dose on the precision of automatic SQUEEZ assessments of RWMA. Chronic myocardial infarction was created by a 2-h occlusion of the left anterior descending coronary artery in 10 swine (heart rates 80–100, ejection fraction 25–57%). CT was performed 5–11 months post infarct using first-pass contrast enhanced segmented cardiac function scans on a 320-detector row scanner at 80 kVp/500 mA. Images were reconstructed at end diastole and end systole with both filtered back projection and using the “standard” adaptive iterative dose reduction (AIDR) algorithm. For each acquisition, 9 lower dose acquisitions were created. End systolic myocardial function maps were calculated using SQUEEZ for all noise levels and contrast-to-noise ratio (CNR) between the left ventricle blood and myocardium was calculated as a measure of image quality. For acquisitions with CNR > 4, SQUEEZ could be estimated with a precision of ± 0.04 (p < 0.001) or 5.7% of its dynamic range. The difference between SQUEEZ values calculated from AIDR and FBP images was not statistically significant. Regional wall motion abnormality can be quantified with good precision from low dose acquisitions, using SQUEEZ, as long as the blood-myocardium CNR stays above 4.
@article{pourmorteza2018precision, doi = {10.1007/s10554-018-1332-2}, url = {https://doi.org/10.1007/s10554-018-1332-2}, year = {2018}, month = mar, publisher = {Springer Science and Business Media {LLC}}, volume = {34}, number = {8}, pages = {1277--1286}, author = {Pourmorteza, Amir and Keller, Noemie and Chen, Richard J. and Lardo, Albert and Halperin, Henry and Chen, Marcus Y. and McVeigh, Elliot}, title = {Precision of Regional Wall Motion Estimates from Ultra-Low-Dose Cardiac {CT} using {SQUEEZ}}, journal = {The International Journal of Cardiovascular Imaging}, demo = {https://www.youtube.com/watch?v=BOXKMkZfpik}, abbr = {pourmorteza2018precision.png} }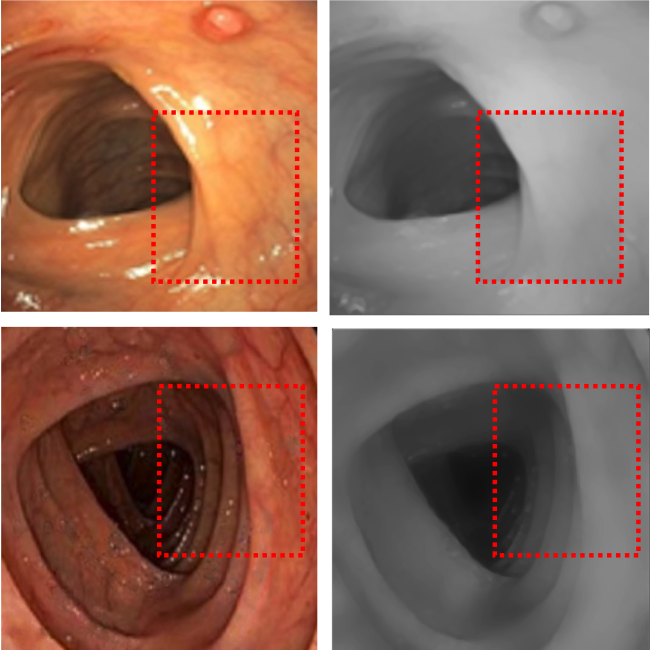 Unsupervised Reverse Domain Adaptation for Synthetic Medical Images via Adversarial TrainingFaisal Mahmood, Richard J. Chen, and Nicholas J. DurrIEEE Transactions on Medical Imaging 2018
Unsupervised Reverse Domain Adaptation for Synthetic Medical Images via Adversarial TrainingFaisal Mahmood, Richard J. Chen, and Nicholas J. DurrIEEE Transactions on Medical Imaging 2018To realize the full potential of deep learning for medical imaging, large annotated datasets are required for training. Such datasets are difficult to acquire due to privacy issues, lack of experts available for annotation, underrepresentation of rare conditions, and poor standardization. The lack of annotated data has been addressed in conventional vision applications using synthetic images refined via unsupervised adversarial training to look like real images. However, this approach is difficult to extend to general medical imaging because of the complex and diverse set of features found in real human tissues. We propose a novel framework that uses a reverse flow, where adversarial training is used to make real medical images more like synthetic images, and clinically-relevant features are preserved via self-regularization. These domain-adapted synthetic-like images can then be accurately interpreted by networks trained on large datasets of synthetic medical images. We implement this approach on the notoriously difficult task of depth-estimation from monocular endoscopy which has a variety of applications in colonoscopy, robotic surgery, and invasive endoscopic procedures. We train a depth estimator on a large data set of synthetic images generated using an accurate forward model of an endoscope and an anatomically-realistic colon. Our analysis demonstrates that the structural similarity of endoscopy depth estimation in a real pig colon predicted from a network trained solely on synthetic data improved by 78.7% by using reverse domain adaptation.
@article{mahmood2018unsupervised, abbr = {mahmood2018unsupervised.png}, doi = {10.1109/tmi.2018.2842767}, url = {https://doi.org/10.1109/tmi.2018.2842767}, year = {2018}, month = dec, publisher = {Institute of Electrical and Electronics Engineers ({IEEE})}, volume = {37}, number = {12}, pages = {2572--2581}, author = {Mahmood, Faisal and Chen, Richard J. and Durr, Nicholas J.}, title = {Unsupervised Reverse Domain Adaptation for Synthetic Medical Images via Adversarial Training}, journal = {{IEEE} Transactions on Medical Imaging}, arxiv = {1711.06606} }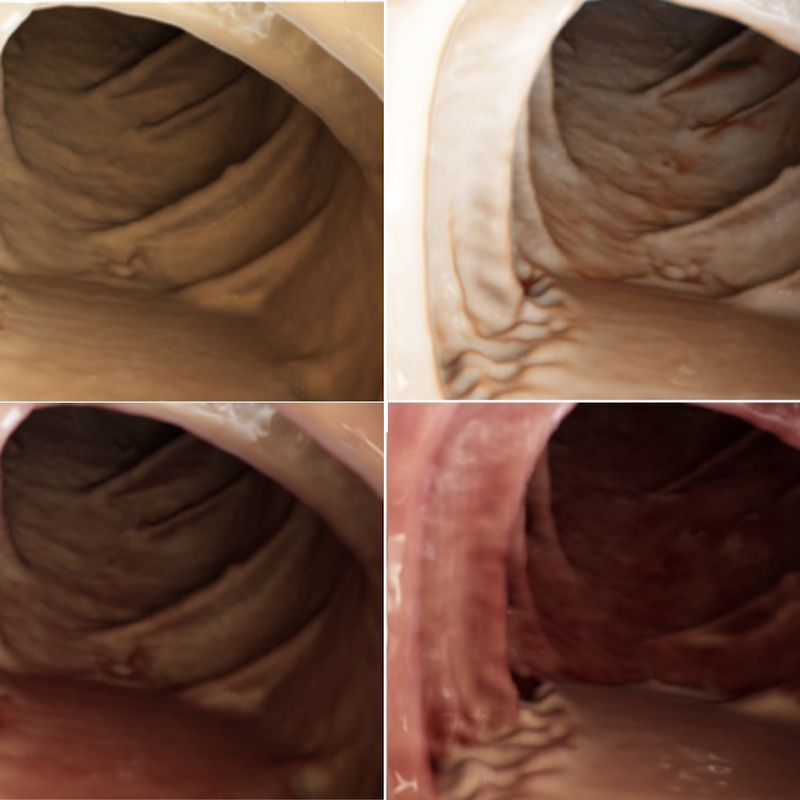 Deep Learning with Cinematic Rendering: Fine-Tuning Deep Neural Networks using Photorealistic Medical ImagesFaisal Mahmood, Richard J. Chen, Sandra Sudarsky, Daphne Yu, and Nicholas J DurrPhysics in Medicine & Biology 2018
Deep Learning with Cinematic Rendering: Fine-Tuning Deep Neural Networks using Photorealistic Medical ImagesFaisal Mahmood, Richard J. Chen, Sandra Sudarsky, Daphne Yu, and Nicholas J DurrPhysics in Medicine & Biology 2018Deep learning has emerged as a powerful artificial intelligence tool to interpret medical images for a growing variety of applications. However, the paucity of medical imaging data with high-quality annotations that is necessary for training such methods ultimately limits their performance. Medical data is challenging to acquire due to privacy issues, shortage of experts available for annotation, limited representation of rare conditions and cost. This problem has previously been addressed by using synthetically generated data. However, networks trained on synthetic data often fail to generalize to real data. Cinematic rendering simulates the propagation and interaction of light passing through tissue models reconstructed from CT data, enabling the generation of photorealistic images. In this paper, we present one of the first applications of cinematic rendering in deep learning, in which we propose to fine-tune synthetic data-driven networks using cinematically rendered CT data for the task of monocular depth estimation in endoscopy. Our experiments demonstrate that: (a) convolutional neural networks (CNNs) trained on synthetic data and fine-tuned on photorealistic cinematically rendered data adapt better to real medical images and demonstrate more robust performance when compared to networks with no fine-tuning, (b) these fine-tuned networks require less training data to converge to an optimal solution, and (c) fine-tuning with data from a variety of photorealistic rendering conditions of the same scene prevents the network from learning patient-specific information and aids in generalizability of the model. Our empirical evaluation demonstrates that networks fine-tuned with cinematically rendered data predict depth with 56.87% less error for rendered endoscopy images and 27.49% less error for real porcine colon endoscopy images.
@article{mahmood2018cinematic, doi = {10.1088/1361-6560/aada93}, url = {https://doi.org/10.1088/1361-6560/aada93}, year = {2018}, month = sep, publisher = {{IOP} Publishing}, volume = {63}, number = {18}, pages = {185012}, author = {Mahmood, Faisal and Chen, Richard J. and Sudarsky, Sandra and Yu, Daphne and Durr, Nicholas J}, title = {Deep Learning with Cinematic Rendering: Fine-Tuning Deep Neural Networks using Photorealistic Medical Images}, journal = {Physics in Medicine {\&} Biology}, arxiv = {1805.08400}, abbr = {mahmood2018cinematic.png}, press = {https://www.auntminnie.com/index.aspx?sec=log&itemID=121635} } Rethinking Monocular Depth Estimation with Adversarial TrainingRichard J. Chen, Faisal Mahmood, Alan Yuille, and Nicholas J DurrarXiv preprint arXiv:1808.07528 2018
Rethinking Monocular Depth Estimation with Adversarial TrainingRichard J. Chen, Faisal Mahmood, Alan Yuille, and Nicholas J DurrarXiv preprint arXiv:1808.07528 2018Monocular depth estimation is an extensively studied computer vision problem with a vast variety of applications. Deep learning-based methods have demonstrated promise for both supervised and unsupervised depth estimation from monocular images. Most existing approaches treat depth estimation as a regression problem with a local pixel-wise loss function. In this work, we innovate beyond existing approaches by using adversarial training to learn a context-aware, non-local loss function. Such an approach penalizes the joint configuration of predicted depth values at the patch-level instead of the pixel-level, which allows networks to incorporate more global information. In this framework, the generator learns a mapping between RGB images and its corresponding depth map, while the discriminator learns to distinguish depth map and RGB pairs from ground truth. This conditional GAN depth estimation framework is stabilized using spectral normalization to prevent mode collapse when learning from diverse datasets. We test this approach using a diverse set of generators that include U-Net and joint CNN-CRF. We benchmark this approach on the NYUv2, Make3D and KITTI datasets, and observe that adversarial training reduces relative error by several fold, achieving state-of-the-art performance.
@article{chen2018rethinking, title = {Rethinking Monocular Depth Estimation with Adversarial Training}, author = {Chen, Richard J. and Mahmood, Faisal and Yuille, Alan and Durr, Nicholas J}, journal = {arXiv preprint arXiv:1808.07528}, year = {2018}, abbr = {chen2018rethinking.png}, arxiv = {1808.07528} }
2016
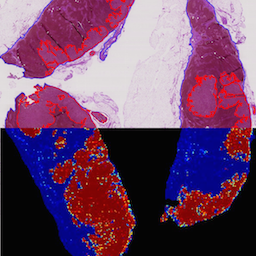 Identifying metastases in sentinel lymph nodes with deep convolutional neural networksRichard Chen, Yating Jing, and Hunter JacksonarXiv preprint arXiv:1608.01658 2016
Identifying metastases in sentinel lymph nodes with deep convolutional neural networksRichard Chen, Yating Jing, and Hunter JacksonarXiv preprint arXiv:1608.01658 2016Metastatic presence in lymph nodes is one of the most important prognostic variables of breast cancer. The current diagnostic procedure for manually reviewing sentinel lymph nodes, however, is very time-consuming and subjective. Pathologists have to manually scan an entire digital whole-slide image (WSI) for regions of metastasis that are sometimes only detectable under high resolution or entirely hidden from the human visual cortex. From October 2015 to April 2016, the International Symposium on Biomedical Imaging (ISBI) held the Camelyon Grand Challenge 2016 to crowd-source ideas and algorithms for automatic detection of lymph node metastasis. Using a generalizable stain normalization technique and the Proscia Pathology Cloud computing platform, we trained a deep convolutional neural network on millions of tissue and tumor image tiles to perform slide-based evaluation on our testing set of whole-slide images images, with a sensitivity of 0.96, specificity of 0.89, and AUC score of 0.90. Our results indicate that our platform can automatically scan any WSI for metastatic regions without institutional calibration to respective stain profiles.
@article{chen2016identifying, title = {Identifying metastases in sentinel lymph nodes with deep convolutional neural networks}, author = {Chen, Richard and Jing, Yating and Jackson, Hunter}, journal = {arXiv preprint arXiv:1608.01658}, year = {2016}, abbr = {chen2016identifying.png} }
2015
 Variations in Glycogen Synthesis in Human Pluripotent Stem Cells with Altered Pluripotent StatesRichard J. Chen, Guofeng Zhang, Susan H. Garfield, Yi-Jun Shi, Kevin G. Chen, Pamela G. Robey, and Richard D. LeapmanPLOS ONE 20152013 Intel STS Semifinalist
Variations in Glycogen Synthesis in Human Pluripotent Stem Cells with Altered Pluripotent StatesRichard J. Chen, Guofeng Zhang, Susan H. Garfield, Yi-Jun Shi, Kevin G. Chen, Pamela G. Robey, and Richard D. LeapmanPLOS ONE 20152013 Intel STS SemifinalistHuman pluripotent stem cells (hPSCs) represent very promising resources for cell-based regenerative medicine. It is essential to determine the biological implications of some fundamental physiological processes (such as glycogen metabolism) in these stem cells. In this report, we employ electron, immunofluorescence microscopy, and biochemical methods to study glycogen synthesis in hPSCs. Our results indicate that there is a high level of glycogen synthesis (0.28 to 0.62 μg/μg proteins) in undifferentiated human embryonic stem cells (hESCs) compared with the glycogen levels (0 to 0.25 μg/μg proteins) reported in human cancer cell lines. Moreover, we found that glycogen synthesis was regulated by bone morphogenetic protein 4 (BMP-4) and the glycogen synthase kinase 3 (GSK-3) pathway. Our observation of glycogen bodies and sustained expression of the pluripotent factor Oct-4 mediated by the potent GSK-3 inhibitor CHIR-99021 reveals an altered pluripotent state in hPSC culture. We further confirmed glycogen variations under different naïve pluripotent cell growth conditions based on the addition of the GSK-3 inhibitor BIO. Our data suggest that primed hPSCs treated with naïve growth conditions acquire altered pluripotent states, similar to those naïve-like hPSCs, with increased glycogen synthesis. Furthermore, we found that suppression of phosphorylated glycogen synthase was an underlying mechanism responsible for altered glycogen synthesis. Thus, our novel findings regarding the dynamic changes in glycogen metabolism provide new markers to assess the energetic and various pluripotent states in hPSCs. The components of glycogen metabolic pathways offer new assays to delineate previously unrecognized properties of hPSCs under different growth conditions.
@article{chen2015variations, doi = {10.1371/journal.pone.0142554}, url = {https://doi.org/10.1371/journal.pone.0142554}, year = {2015}, month = nov, publisher = {Public Library of Science ({PLoS})}, volume = {10}, number = {11}, pages = {e0142554}, author = {Chen, Richard J. and Zhang, Guofeng and Garfield, Susan H. and Shi, Yi-Jun and Chen, Kevin G. and Robey, Pamela G. and Leapman, Richard D.}, editor = {Lako, Majlinda}, title = {Variations in Glycogen Synthesis in Human Pluripotent Stem Cells with Altered Pluripotent States}, journal = {{PLOS} {ONE}}, abbr = {chen2015variations.png}, honor = {2013 Intel STS Semifinalist} }